Development of Ultra-rapid PCR System for Genotyping of Sacbrood Virus
Abstract
Sacbrood virus (SBV) is one of the main honeybee pathogens. It has been detected worldwide. Five genotypes of SBV were proposed based on the analysis of coding DNA sequences (CDSs) and deduced amino acids. However, a reliable tool to examine each SBV-genotype has not been developed yet. To establish a PCR system for SBV-genotyping, designation of specific primer-pairs and construction of standard DNAs for 5 genotypes were carried out. The detection system for SBV-genotypes was optimized using each standard DNA and specific primer-pairs. Then PCR system for SBV-genotyping was applied with SBV-infected honeybee samples. Interestingly, in some of SBV-infected Apis mellifera, 2134D51 genotype (kSBV) and 2100D0 genotype (wSBV) were both detected, however, 2100D0 was existed dominantly. Whereas, 2100D0 was not detected from SBV-infected samples of Apis cerana, only 2134D51 was identified as solo pathogen in this species. PCR system for SBV-genotyping might be helpful to understand SBV-distribution and spreading.
Keywords:
Sacbrood virus, Genotype, Ultra-Rapid PCR, Apis cerana, Apis melliferaINTRODUCTION
Sacbrood virus (SBV), one of the main pathogenic viruses in the honeybee, belongs to family Iflaviridae, genus Iflavirus. SBV has a circular capsid 28nm in diameter. There is only one open reading frame in the SBV genome, which encodes a polyprotein consisting of 2858 amino acids (Ghosh et al., 1999). SBV causes the failure of larval pupation in honeybee, color of infected-larva changes from white to pale yellow followed by death (Bailey, 1975).
SBV was first recorded in 1913, but was not characterized until 1964 (White, 1917; Bailey et al., 1964). It has been found worldwide, including North America (van Engelsdorp et al., 2009), South America (Freiberg et al., 2012), Europe (Grabensteiner et al., 2001; Tentcheva et al., 2004), Australia (Anderson and Gibbs, 1988), South Africa (Grabensteiner et al., 2001), and Asia (Zhang et al., 2001; Choi et al., 2010; Ma et al., 2013; Nguyen and Le, 2013).
After many geographical variants of SBV have been detected, these regional SBV variants were called as common name, such as cSBV (Chinese variant; Mingxiao et al., 2011), kSBV (SBV in Korea; Choi et al., 2010; Choe et al., 2012), vSBV (SBV in Vietnam; Nguyen and Le, 2013), iSBV (SBV in India; Kshirsagar et al., 1982; Rao et al., 2016), and tSBV (SBV in Thailand). In contrast. the first isolated SBV group from western honeybee Apis mellifera, that has been already worldwide detected from A. mellifera, was called simply as wSBV (western SBV; Lee et al., 2017).
Based on amino acid sequences of SBVs, Lee et al. (2017) found a characteristic deletion, located between two capsid genes, VP1 and VP3. Using position and nucleotides of this deletion, designated 2100D, five SBV genotypes were proposed, such as 2134D51, 2119D39 2119D30, 2100D0 and 2134D3. These genotypes were also well matched to common names of SBV, such as kSBV and vSBV, cSBV, iSBV, wSBV, and English SBV (eSBV), respectively.
The origin of these SBV genotypes and whether the variant in one geographical region is able to infect honeybees in another region is an interesting and unresolved issue. Furthermore, the characteristics of each genotype are still unclear, and a tool for fast detection and accurate differentiation of these geographical SBV variants has not been developed.
SBV can persistently exist in honeybee colony without clear symptoms in adult bee, particularly when the virus accumulated in brains of infected bees (Bailey and Fernando, 1972). Hence, besides the observation of symptoms in infected honeybees, the molecular detection by reverse transcription-PCR (RT-PCR) has been an accurate and rapid means of virus detection when the genomic nucleotide sequences of SBV have been determined (Grabensteiner et al., 2001).
A PCR detection method for kSBV was developed in 2008 by Nguyen Thi et al. (2009). Then SBV detection using ultra-rapid real-time PCR was developed by Yoo et al. (2012) for detection result within 22 minutes including reverse transcription step. This SBV detection method has been greatly improved for rapid detection, for instance, an ultra-rapid RT-PCR against kSBV that can detect the virus within 6 minutes 12 seconds (Min et al., 2016).
This study was conducted to develop a method for accurate and fast determination of SBV genotypes using ultra-rapid real-time PCR, it was expected to be the most rapid method of genotype detection. It plays an important role in establishing further research on the origin and infection patterns of each genotype by which a honey bee variant with resistance to SBV might be found.
MATERIALS AND METHODS
SBV-infected honeybee specimens
Adult bees from the colonies with symptoms of SBV infection in larvae were collected in South Korea. Samples for SBV genotyping consist of two Apis mellifera samples designated as S2 and S3 which were collected in Yongin (2017), other four A. cerana samples were designated as S1, S4, S5, and S6 in which S1 was collected in Chungju (2013), S4 and S6 were from Okcheon (2014), and S5 was from Goyang (2014). Samples for standard DNA construction were collected in Okcheon (2014) and Seongju (2017) (Fig. 1).
Total RNA extraction from honeybee samples
Total RNA from infected adult bees was extracted using RNAiso Plus reagent (Takara, Japan), the procedure was based on the manufacturer’s instruction. The precipitated and dried RNA was dissolved in 100μl of RNase-free water. The concentration of the extracted RNA was determined using a biophotometer (Eppendorf, Germany), it was then diluted into 100ng/μl and used for SBV detection by RT-PCR.
Primer design for SBV detection and genotype identification
Two universal primer pairs were designed to detect all SBV strains: SBVD-F1/SBVD-R1 and SBVD-F2/SBVDR2 (Table 1). A DNA fragment 582 bp long was amplified from CDS of genotype 2100D0 by primer pair SBVD-F1/SBVD-R1. Size of amplicon in other genotypes were 579 bp (2134D3), 552 bp (2119D30), 543 bp (2119D39), and 531 bp long (2134D51) (Fig. 2).
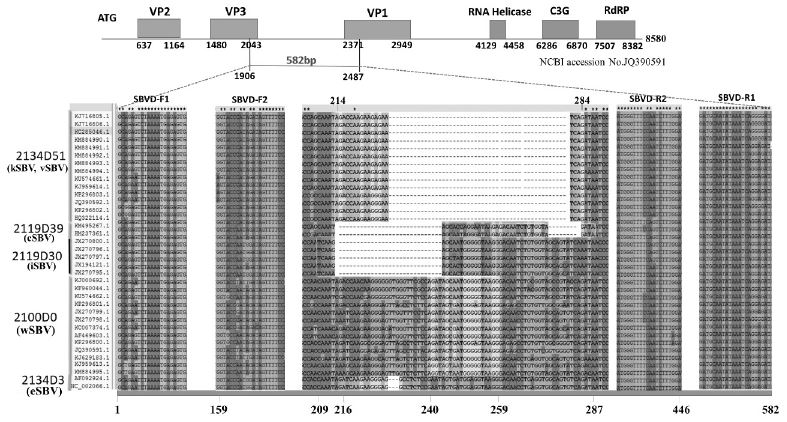
The alignment of SBV genomes shows various numbers of missing nucleotide among the 5 genotypes from which the specific genotyping primers were designed. The alignment shows 51, 39, 30, and 3 nucleotides were deleted in the missing gap of genotype 2134D51 (kSBV and vSBV), 2119D39 (cSBV), 2119D30 (iSBV), 2134D3 (eSBV), respectively. These deleting nucleotides were identified based on the comparison with genotype 2100D0 (wSBV). Specific primer of each genotype was selected containing the deleting gap in order to reduce the non-specific amplification. Universal primer pairs SBVD-F1/SBVD-R1 and SBVD-F2/SBVD-R2 were designed to amplified a fragment 582 bp (1906-2487) and 288 bp long (2065-2352), respectively, on CDS of 2100D0 genotype. In the figure, only nucleotides at primer designing and deleting positions are shown. This figure were created based on the analysis of Lee et al. (2017).
The second universal primer pair, SBVD-F2/SBVD-R2, amplified a segment from the positions 159 to 446 (288 bp) on the 582 bp fragment of 2100D0. This primer pair was designed to combine with genotyping primers for SBV genotype identification (Fig. 3).
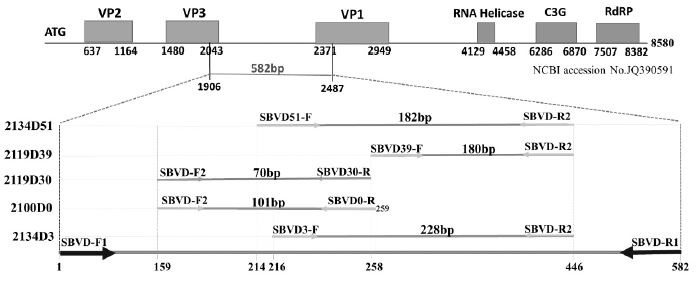
Schematic diagram shows primer pairs and amplicon size of genotyping detection. Primer pairs for genotyping detection were SBVD51-F/SBVD-R2 (2134D51), SBVD39-F/SBVD-R2 (2119D39), SBVD30-R/SBVD-F2 (2119D30), SBVD0-R/SBVD-F2 (2100D0), SBVD3-F/SBVD-R2 (2134D3). Their target size was 182 bp, 180 bp, 70 bp, 101 bp, 235 bp and 226 bp long, respectively.
Primers for specific genotype detection were designed based on the result of nucleotide alignment. Each genotyping primer was selected at the position that contained the missing gap. By this method the non-specific amplification can be avoided. Five specific genotyping primers were designed to enable the accurate differentiation of 5 SBV genotypes from infected honeybee samples (Table 1).
Construction of genotyping standard DNA
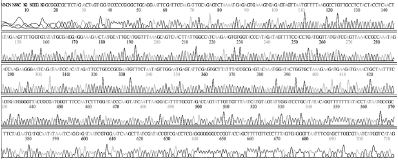
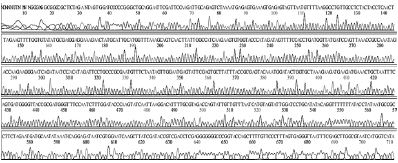
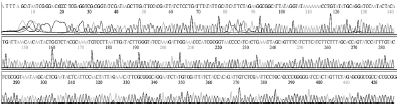
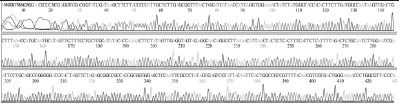
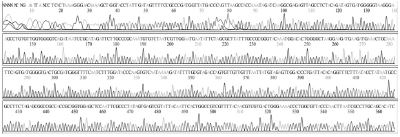
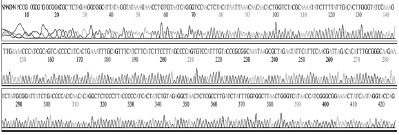
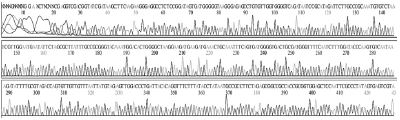
Standard DNAs for each genotype were constructed, these DNAs were used as positive controls in ultra-rapid PCR. The successful construction of these DNAs was confirmed by two directions sequencing (SolGent, Korea) (Table S1).
The 531 bp long standard fragment of genotype 2134D51 was amplified using primer pair SBVD-F1/SBVD-R1 from SBV-infected honeybee sample (A. cerana) collected in Okcheon, South Korea (2014). This fragment was inserted in the pBlueXcm vector by the TA cloning method using restriction enzyme XcmI and T4 ligase (New England Biolabs, USA). The recombinant vector was designated as pSBVD51.
Standard DNA for genotype 2100D0 was also derived from SBV-infected honeybee sample (A. mellifera) collected in Seongju, South Korea (2017). A 582 bp fragment was amplified using universal primer pair SBVD-F1/SBVD-R1. This fragment was cut by restriction enzymes KpnI (New England BioLabs,) and XbaI (Fermentas, USA), resulting in a 400 bp long fragment, from position 159 to 558. The digestion fragment was inserted in vector pBlueXcm cut by the same restriction enzymes. A recombinant vector carries the standard DNA of genotype 2100D0 was constructed and designated as pSBVD0.
Standard DNAs of the remaining genotypes (2119D30, 2119D39, and 2134D3) were constructed containing a part for detection of specific genotyping primer plus another part derived from standard DNA of genotype 2100D0 or 2134D51 (Table S2 and Fig. S1-3).
Reverse transcription and polymerase chain reaction
SBV was detected by RT-PCR using universal primer pair SBVD-F1/SBVD-R1 and 100ng of total RNA that was isolated from infected honeybee samples. The RT-PCR conditions were: 5 min at 50°C for RT, followed by PCR at 95°C (30s) and 45 cycles of 95°C (4s)-51°C (4s)-72°C (5s). Genotyping PCR was performed at 95°C (30s) of pre-denaturation and followed by 35 cycles of 95°C(3s)-54°C (2s)-72°C (2s). PCR condition for sensitive inspection of genotyping primers was performed at the same condition as genotyping PCR except the number of cycle was 50 cycles instead of 35 cycles. GENECHECKERTM Ultra-Rapid Real-time PCR System and reagent 2X Rapi RT Master Mix (Gen-esystem Co., Ltd., Korea) were used.
To construct standard DNA for genotype 2134D51 and 2100D0, total RNA (100ng) was used in conventional RT-PCR. The RT step was 50°C for 10 min with the oligo dT primer, followed by 40 cycles of PCR amplification at 95°C for 5 min, followed by 40 cycles of 95°C (30s)-51°C (30s)-72°C (30s), and 7 min post-polymerization at 72°C.
SBV genotype identification
Each genotyping primer pair was separately inspected on standard DNAs of 5 genotypes using ultra-rapid real-time PCR to confirm the specific amplification. Additionally, the mixture consisting standard DNA of 5 genotypes (106 molecules of each genotype) was utilized for each genotyping primer pair to examine the specific detection.
For SBV genotype identification from infected honeybee samples, SBV was initially detected by ultra-rapid RT-PCR using universal primer pair SBVD-F1/SBVD-R1. The PCR product of positive detection was 1/100 diluted and used for genotyping identification by specific genotyping primers. Genotyping primer pairs and amplicon size were shown in Fig. 3.
RESULTS
Standard DNA for SBV genotyping
The standard DNAs of 5 genotypes were constructed. These DNAs were used as positive control for genotyping identification. The position of these fragments, corresponding to position at the 582 bp long fragment of 2100D0 amplified by primer pair SBVD-F1/SBVD-R1, were 1 to 582 (2134D51), 258 to 582 (2119D39), 1 to 258 (2119D30), 218 to 558 (2134D3), 159 to 558 (2100D0). The number of deleting nucleotide in each fragment was illustrated inside the specific genotype primer. The size (in bp) of these DNAs were 531 (2134D51), 316 (2119D39), 228 (2119D30), 338 (2134D3), and 400 (2100D0) (Fig. 4).
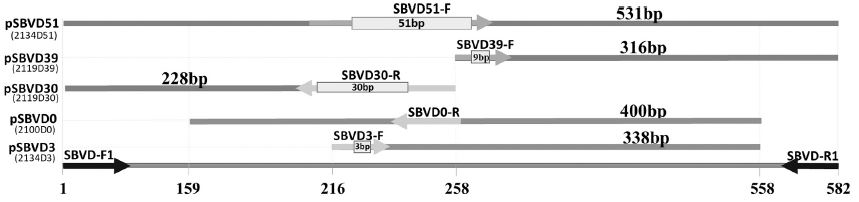
Standard DNAs of 5 SBV genotypes which were used as positive control for genotyping PCR. Standard DNA of genotype 2134D51, 2119D39, 2119D30, 2100D0, and 2134D3 was 531 bp, 316 bp, 228 bp, 400 bp, and 338 bp long, respectively. The position of each fragment was indicated based on the length (582 bp) of fragment amplified by universal primer SBVD-F1/R1 in genotype 2100D0, was indicated. Direction and position of specific genotyping primer was illustrated in each standard DNA. Number of deleting nucleotide was also indicated in each genotyping primer.
The recombinant vector carrying each standard fragment was designated as pSBVD51 (2134D51), pSBVD39 (2119D39), pSBVD30 (2119D30), pSBVD0 (2100D0), and pSBVD3-338 (2134D3).
Accuracy of SBV genotyping on standard DNA
The accuracy of genotyping detection was demonstrated by individually inspecting each genotyping primer pair on standard DNAs of 5 genotypes in ultra-rapid real-time PCR. The fluorescent curves and Tm value showed the specific detection on standard DNA. Each genotyping primer amplified only its standard DNA (Fig. 5), there was no non-specific amplification within 35 cycles of PCR. Furthermore, in the mixture of all genotype DNAs that mimic the presence of various genotypes in one infected sample. The accurate detection of each genotyping primer pair on its standard DNA was acquired. The result was determined based on similarity of Tm value of amplification in the mixture DNA and single genotype DNA used in each primer pair (Fig. 5).
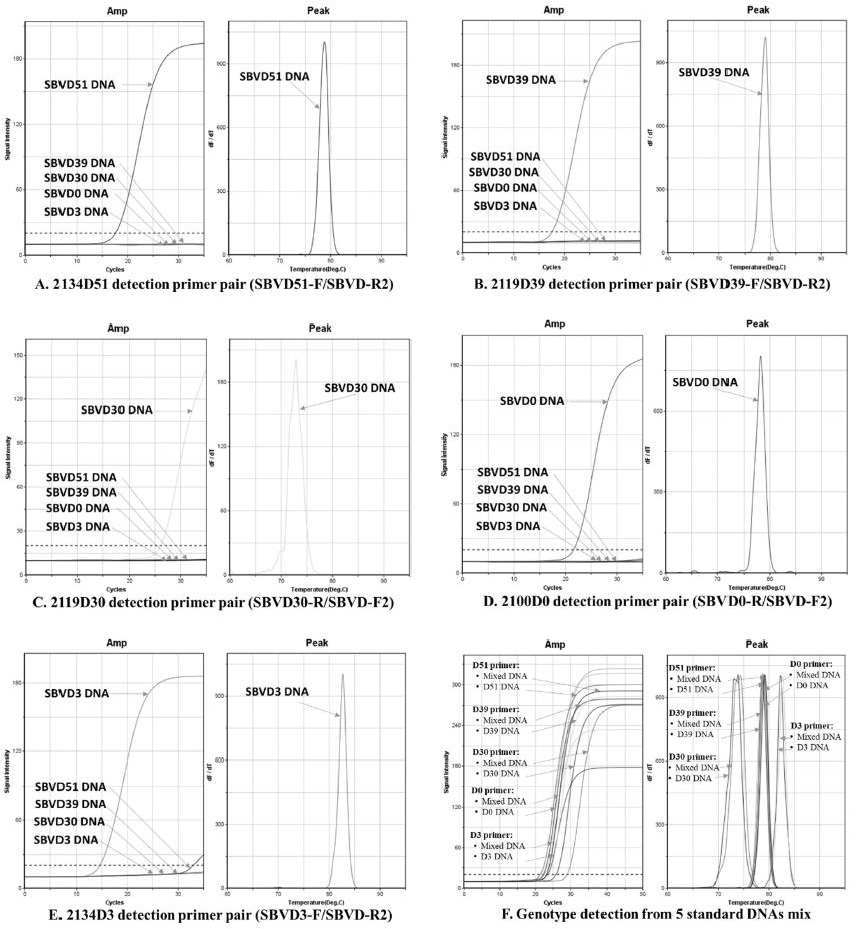
Fluorescent curves of genotyping PCR that was performed using standard DNAs and specific genotyping primers. Five standard DNAs were separately used as DNA template for each genotyping primer pair in each PCR performance (A-E), the genotyping primer accurately detected its standard DNA within 35 cycles PCR performance, the non-specific amplification was not seen in the graph of Tm value in each primer pair. Genotyping primer was also able to amplify only its standard DNA in the mixture of 5 standard DNAs, the accurate amplification was achieved by the similar Tm curve of amplification from DNA mix and from single genotype DNA.
The accurate detection of genotyping primers on its standard DNAs demonstrated that these genotyping primers can be used for genotyping identification from infected honeybee sample.
Sensitivity of genotyping primers
Standard DNA of each genotype was serially diluted from the concentration of 108 [2.64x108 (2134D51); 2.84x108 (2119D39); 2.91x108 (2119D30); 2.83x108 (2100D0); 2.84x108 (2134D3)] to 100 molecule/μl, and 1μl of each concentration was used to evaluate the sensitivity of each genotyping primer pair. Primer pair SBVD3-F/SBVD-R2 (2134D3) was less sensitive than other primers with limit detection was 2.84x102 copies of DNA template. The most sensitive primer pair was SBVD30-R/SBVD-F2 (2119D30), it was able to detect the standard DNA to 2.91x100 copies. The limit detection of other primer pairs, SBVD51-F/SBVD-R2 (2134D51), SBVD39-F/SBVD-R2 (2119D39), SBVD0-R/SBVD-F2 (2100D0), was 2.64x101, 2.84x101 and 2.83x101 copies of standard DNA, respectively (Fig. 6).
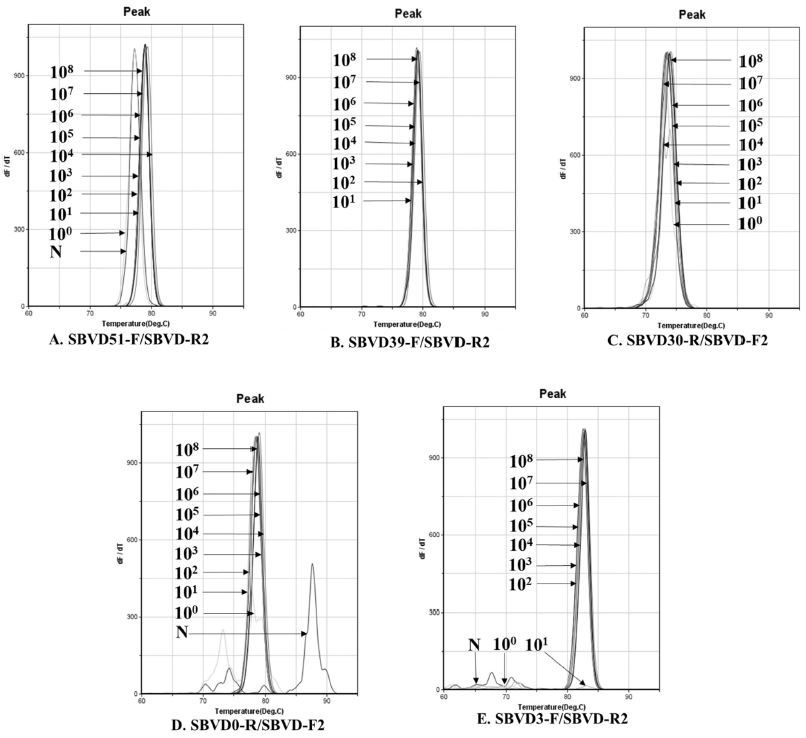
Fluorescent curves of melting temperature in ultra-rapid real-time PCR that was performed to inspect the sensitivity of genotyping primers. The specific amplification by each genotyping primer pair was determined based on the similarity of fluorescent curves of melting temperature. The minimum detection of primer pair SBVD51-F/SBVD-R2 (2134D51, A), SBVD39-F/SBVD-R2 (2119D39, B), SBVD30-R/SBVD-F2 (2119D30, C), SBVD0-R/SBVD-F2 (2100D0, D), and SBVD3-F/SBVD-R2 (2134D3, E) was 101, 101, 100, 101, and 102 copies of DNA template, respectively.
The regression equation of molecule number and Ct value was established, it was y=-3.017x+40.834, R2=0.9798; y=-3.6374x+42.376, R2=0.9873; y=-3.2115x+43.403, R2=0.9912; y=-2.9578x+40.526, R2=0.9925; y=-3.5146x+41.673, R2=0.9557 in primer pair of genotype 2134D51, 2119D39, 2119D30, 2100D0 and 2134D3, respectively.
SBV genotyping from SBV-infected honeybee samples
SBV was detected from total RNA of 6 honeybee samples by universal primer pair SBVD-F1/SBVD-R1 in ultra-rapid RT-PCR. The Tm values in A. cerana samples (S1, S4, S5, S6) were approximately similar to the value of 2134D51 standard DNA, whereas the Tm value in A. mellifera samples (S2 and S3) was higher than it of 2134D51 standard DNA (Fig. 7A). Additionally, electrophoresis showed the bands approximately 582 bp and 531 bp long in A. mellifera and A. cerana samples, respectively (Fig. 7B).
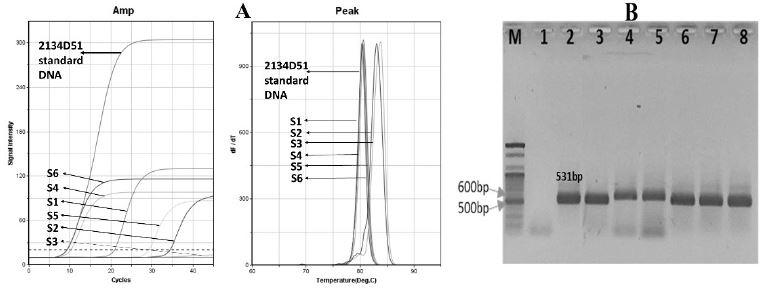
Fluorescent curves of SBV detection RT-PCR and electrophoresis showed the positive detection of SBV in 6 infected honeybee samples using universal primer pair SBVD-F1/SBVD-R1. The graph (A) and agarose gel image (B) demonstrated the positive detection of SBV in 6 honey bee samples. Four A. cerana samples S1, S4, S5, and S6 had Tm value (A) and a band 531 bp long (B, lane 3, 6, 7, 8) in electrophoresis similar to 2134D51 standard DNA (B, lane 2). The Tm values in A. mellifera samples (S2, S3) was higher than it of 2134D51 standard DNA (A), and the band around 582 bp long was observed in agarose gel (B, lane 4, 5). Lane M and 1 denote the 100-bp marker and negative control (without DNA template), respectively.
Therefore, SBV in A. cerana samples might belong to genotype 2134D51 and other genotypes were present in A. mellifera samples. This universal primer pair is impossible to identify the genotype due to the length PCR product among the genotypes are not greatly different.
The PCR products of positive detection were diluted 100 times and used for genotyping PCR. In each genotype detection primer, the standard DNA was adjacent to the sample DNA template in PCR. The positive detection was determined by comparing the Tm value of sample DNA to standard DNA, observing the melting curves, then the amplicon size was confirmed in electrophoresis. Finally, the sequences of genotype detection was compare to the SBV sequences deposited on NCBI using Nucleotide blast tool. Genotype 2134D51 and 2100D0 were both detected in 2 A. mellifera samples (S2, 3), whereas only genotype 2134D51 was existed in 4 A. cerana samples (S1, 4, 5, 6), it was consistent with the report of Choi et al. (2010) that the 2134D51 variant (kSBV) caused the collapse of over 300,000 populations of A. cerana in South Korea in 2009.
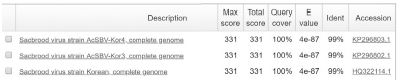
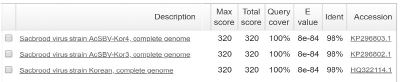
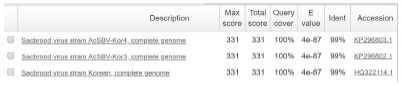
The Tm value of genotype 2134D51 and 2100D0 detection from sample DNAs and standard DNAs showed the approximate Tm value. The Tm value of sample DNA/standard DNA from 2134D51 detection primer pair (SBVD51-F/SBVD-R2) was 79.44/79.12°C in 4 A. cerana samples, and 79.12/78.47°C in 2 A. mellifera samples. The detection was confirmed in electrophoresis by a band 182-bp long was observed in all samples. Furthermore, the sequencing result of the 6 amplicons demonstrated the accurate genotyping identification. These sequences showed the highest homology to sequences of 2134D51 genotype (kSBV) with accession numbers on NCBI are KP296803, KP296802, and HQ322114 ( Table S3).
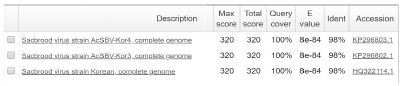
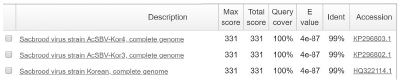
The positive detection of SBV genotype 2100D0 was observed only in A. mellifera samples, the Tm value of standard DNA/sample DNA was 79.12/79.77 (S2), 79.12/79.77 (S3). The 101 bp long band of amplicon was confirmed in electrophoresis. Sequencing result showed the highest identity with 2100D0 genotype strains isolated from A. mellifera host that originated from South Korea and Australia, NCBI accession number are MF623170, JQ390591, KY887699, KP296800 (Table S4).
Other genotypes, 2119D39, 2119D30, and 2134D3 were negative detection in all samples (Fig. 8). Total time required for genotype detection using the RT-PCR system was 38 minutes 23 seconds, in which 24 minutes and 22 seconds for SBV detection RT-PCR and 14 minutes 01 second to finish 35 cycles of genotype identification PCR. The accuracy of genotyping identification from honeybee sample showed a tool for geographical SBV variants identification was established.
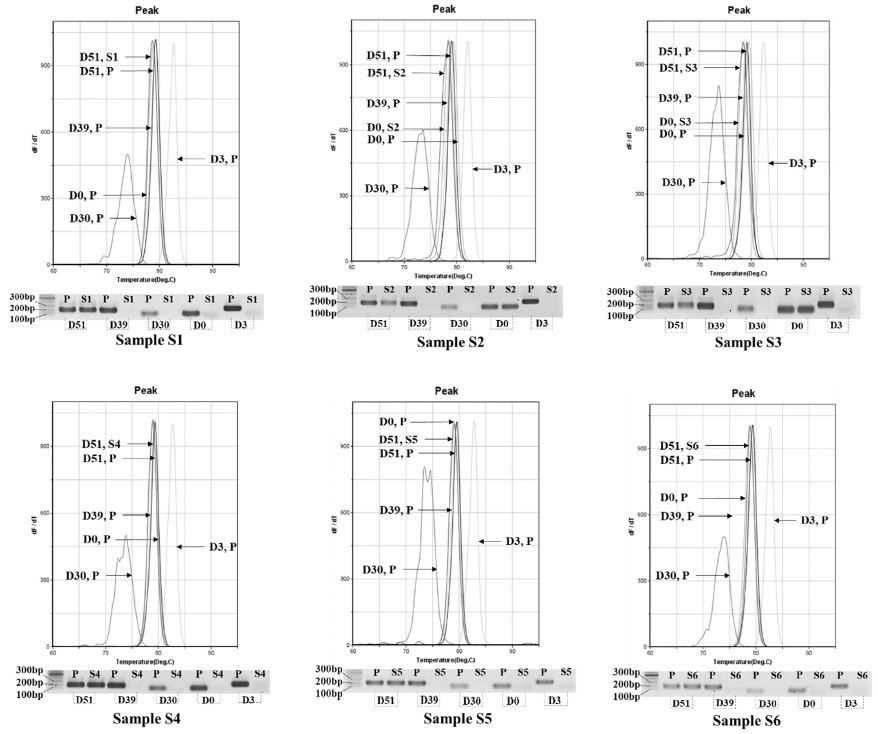
Fluorescent curves of melting temperature in genotyping PCR and electrophoresis show the positive detection of SBV genotypes in 6 honeybee samples. The diluted DNA of SBV from 6 honeybee samples was individually used for SBV genotype identification. DNA of each sample was utilized as template for 5 genotyping primer pairs in 10 wells of PCR chip in each PCR performance. Primer pairs for genotype 2134D51, 2119D39, 2119D30, 2100D0, 2134D3 were denoted as D51, D39, D30, D0, and D3, respectively. In the graph of fluorescent curves sample DNAs of sample S1-6 was denoted as S1-S6, and standard DNA was “P”. Only the positive detection from sample DNA and standard DNA template was seen in the graphs. Fluorescent curves of Tm value showed genotype 2134D51 were detected in 6 samples and an expected band 182 bp long (D51) was seen in agarose gel. The positive detection of genotype 2100D0 was observed only in A. mellifera samples (S2, 3) with a band 101 bp long in agarose gel. Other genotypes, 2119D39, 2119D30, and 2134D3 were negative detection.
In spite of the fact that two SBV genotypes (2134D51 and 2100D0) co-invaded in A. mellifera samples, the molecule number of 2100D0 genotype was markedly higher than it of 2134D51 genotype. Based on the regression equation of Ct value and molecule number of 2134D51 detection primer (y=-3.017x+40.834) and 2100D0 primer (y=-2.9578x+40.526), the molecule number of genotype 2134D51 was 2.08x105 (S2), and 2.02x105 (S3) copies, whereas number of molecule of 2100D0 genotype was 4.14x108 and 1.71x109 copies in the two samples, respectively. This detection was consistent to the study of Gong et al. (2016) that the SBV isolated from A. cerana was able to infect A. mellifera with low prevalence and pathogenicity.
DISCUSSION
The SBV genotype was accurately identified by nested PCR using universal primer pair and followed by genotyping primer. The detection could be performed using a genotyping primer directly on total RNA extracted from honeybee samples. However, the use of many primers on various DNA templates could lead to the amplification of many non-specific positions on SBV genome as well as on RNA originating from the honeybee. Such non-specific amplification might result in inaccurate identification.
Furthermore, the nested PCR is possible to improve the sensitivity of detection as the number of SBV DNA was amplified by universal primer pair, then the genotyping PCR was expected to have more accurate detection. Therefore, the nested PCR method was applied in this study by 2 PCR performances. Although it takes more time spending, the result is outweighed. The PCR system from this study can be utilized to select the desired SBV genotype in studies of the infecting ability among the genotypes or the relationship of the SBV genotypes. With these understandings, a strategy for SBV prevention might be established.
The result in this study demonstrated the presence of 2 genotypes (2134D51 and 2100D0) in A. mellifera host, but only genotype 2134D51 was present in A. cerana. The role of 2134D51 variant was well known in South Korea that have great contribution to the collapse of A. cerana, and the SBV variant 2100D0 is worldwide pathogenic virus of A. mellifera. However, the influence of genotype 2134D51 in A. mellifera with low number of copy has not been well known.
The high variation in sequence of SBV genotype 2100D0 was detected in one A. mellifera colony (Truong et al., 2017). Therefore, there could be possible explanation for this phenomenon is that when genotype 2100D0 infected A. mellifera, the mutation occurred in genotype 2100D0 resulting in genotype 2134D51, this variant was then released from A. mellifera and pose a threat to A. cerana.
To figure out the answer for this hypothesis, further research needs to be conducted, such as the artificial infection of each SBV genotype in separately honeybee species combining with more observation and molecular analysis. Additionally, the genome of SBV variants in different countries had various number of deleted nucleotides at the same genome position when compared to genome of 2100D0 variant, this mutation could allow such SBV variant to adapt to the geographical honey bee variant. Therefore, the hybridization among those geographical honeybee strains could be one of the methods to detect a honeybee variant resistant to the local SBV genotype.
An ultra-rapid PCR system with specific genotyping primers and standard DNAs were successfully established for accurate identification of SBV genotype. This should be a useful tool for further research on the relationship of SBV genotypes on a worldwide scale, and in studies of infection pattern of each genotype. However, in this study the honeybee samples with SBV genotypes distributed in China, India, and the United Kingdom have not been tested due to the lack of these samples.
Acknowledgments
This work was supported by Business for Cooperative R&D between Industry, Academy, and Research Institute funded Korea Small and Medium Business Administration in 2017 (Grant No. C0563751). This work was also supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries (IPET) through Advanced Production Technology Development Program (115102-03) and Kyonggi University’s Graduate Research Assistantship 2018.
References
-
Anderson, D. L., and A. J. Gibbs, (1988), Inapparent virus infections and their interactions in the pupae of the honey bee (Apis mellifera Linnaeus) in Australia, Journal of General Virology, 69, p1617-1625.
[https://doi.org/10.1099/0022-1317-69-7-1617]

-
Bailey, L., A. J. Gibbs, and R. D. Woods, (1964), Sacbrood virus of the larval honey bee (Apis mellifera Linnaeus), Virology, 23, p425-429.
[https://doi.org/10.1016/0042-6822(64)90266-1]

-
Bailey, L., and E. F. W. Fernando, (1972), Effects of sacbrood virus on adult honey-bees, Annals of Applied Biology, 72, p27-35.
[https://doi.org/10.1111/j.1744-7348.1972.tb01268.x]

-
Bailey, L., (1975), Recent research on honey bee viruses, Bee World, 56, p55-64.
[https://doi.org/10.1080/0005772x.1975.11097544]

-
Choe, S. E., L. T. Nguyen, J. H. Noh, C. H. Kweon, K. E. Reddy, H. B. Koh, K. Y. Chang, and S. W. Kang, (2012), Analysis of the complete genome sequence of two Korean sacbrood viruses in the honey bee, Apis mellifera, Virology, 432, p155-161.
[https://doi.org/10.1016/j.virol.2012.06.008]

- Choi, Y. S., M. Y. Lee, I. P. Hong, N. S. Kim, H. K. Kim, K. G. Lee, and M. L. Lee, (2010), Occurrence of sacbrood virus in Korean Apiaries from Apis cerana (Hymenoptera: Apidae), Korean Journal of Apiculture, 25(3), p187-191.
-
Freiberg, M., D. D. Jong, D. Message, and D. Cox-Foster, (2012), First report of sacbrood virus in honey bee (Apis mellifera) colonies in Brazil, Genetic and Molecular Research, 11(3), p3310-3314.
[https://doi.org/10.4238/2012.September.12.14]

-
Ghosh, R. C., B. V. Ball, M. M. Willcocks, and M. J. Carter, (1999), The nucleotide sequence of sacbrood virus of the honeybee: an insect picornavirus, Journal of General Virology, 80, p1541-1549.
[https://doi.org/10.1099/0022-1317-80-6-1541]

-
Gong, H. R., X. X. Chen, Y. P. Chen, F. L. Hu, J. L. Zhang, Z. G. Lin, J. W. Yu, and H. Q. Zheng, (2016), Evidence of Apis cerana Sacbrood virus infection in Apis mellifera, Applied and Environmental Microbiology, 82, p2256-2262.
[https://doi.org/10.1128/AEM.03292-15]

-
Grabensteiner, E., W. Ritter, M. J. Carter, S. Davison, H. Pechhacker, J. Kolodziejek, O. Boecking, I. Derakhshifar, R. Moosbeckhofer, E. Licek, and N. Nowotny, (2001), Sacbrood virus of the honeybee (Apis mellifera): rapid identification and phylogenetic analysis using reverse transcription-PCR, Clinical and Diagnostic Laboratory Immunology, 8(1), p93-104.
[https://doi.org/10.1128/CDLI.8.1.93-104.2001]

- Kshirsagar, K. K., U. C. Saxena, and R. M. Chauhan, (1982), Occurrence of sacbrood disease in Apis cerana indica F, Indian Bee Journal, 44, p8-9.
-
Lee, C. W., M. S. Yoo, S. J. Lim, J. M. Kim, Y. S. Cho, and B. S. Yoon, (2017), A proposal on the new genotyping of Sacbrood viruses for the definition of Korean Sacbrood virus (kSBV), Korean Journal of Apiculture, 32(2), p89-97.
[https://doi.org/10.17519/apiculture.2017.06.32.2.89]

-
Ma, M. X., Y. N. Yin, X. L. Xu, L. Zhang, Y. F. Li, and Z. D. Luan, (2013), Genetic characterization of VP1 gene of seven Sacbrood virus isolated from three provinces in northern China during the years 2008-2012, Virus Research, 176, p78-82.
[https://doi.org/10.1016/j.virusres.201304.018]

-
Min, S. H., J. H. Wang, S. J. Lim, C. W. Lee, and B. S. Yoon, (2016), The Most Rapid Detection Method against Korean Sacbrood Virus using Ultra-Rapid Reverse-Transcription Real-Time PCR (URRTRT-PCR), Korean Journal of Apiculture, 31(2), p121-131.
[https://doi.org/10.17519/apiculture.2016.06.31.2.121]

-
Mingxiao, M., L. Ming, C. Jian, Y. Song, W. Shude, and L. Pengfei, (2011), Molecular and Biological Characterization of Chinese Sacbrood Virus LN Isolate, Comparative and Functional Genomics, 2011, 409386.
[https://doi.org/10.1155/2011/409386]

-
Nguyen, N. T. B., and T. H. Le, (2013), Complete genome sequence of sacbrood virus strain SBM2, isolated from the honeybee Apis cerana in Vietnam, Genome Announcements, 1(1), e00076-12.
[https://doi.org/10.1128/genomea.00076-12]

- Nguyen Thi, K. C., M. S. Yoo, M. H. Kang, S. H. Han, C. H. Yun, B. S. Yoon, (2009), Development of Real-time PCR Assay for the Detection of Sacbrood Virus in Honeybee (Apis mellifera L.), Korean Journal of Apiculture, 24(1), p15-21.
-
Rao, K. M., S. Katna, B. S. Rana, and R. Rana, (2016), Thai sacbrood and sacbrood viruses versus European foulbrood of hive bees in India - a review, Journal of Apicultural Research, 54(3), p192-199.
[https://doi.org/10.1080/00218839.2016.1145417]

-
Tentcheva, D., L. Gauthier, N. Zappulla, B. Dainat, F. Cousserans, M. E. Colin, and M. Bergoin, (2004), Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France, Applied and Environmental Microbiology, 70(12), p7185-7191.
[https://doi.org/10.1128/AEM.70.12.7185-7191.2004]

-
Truong, A. T., J. M. Kim, S. J. Lim, M. S. Yoo, Y. S. Cho, and B. S. Yoon, (2017), High Level of Sequence-Variation in Sacbrood Virus (SBV) from Apis mellifera, Korean Journal of Apiculture, 32(4), p281-293.
[https://doi.org/10.17519/apiculture.2017.11.32.4.281]

-
vanEngelsdorp, D., J. D. Evans, C. Saegerman, C. Mullin, E. Haubruge, B. K. Nguyen, M. Frazier, J. Frazier, D. Cox-Foster, Y. Chen, R. Underwood, R. D. Tarpy, and J. S. Pettis, (2009), Colony collapse disorder: a descriptive study, PLoS One, 4, e6481.
[https://doi.org/10.1371/journal.pone.0006481]

- White, G. F., (1917), Sacbrood, In Bulletin no, 431, p1-55, United States Department of Agriculture.
-
Yoo, M. S., K. C. Thi, N. P. Van, S. H. Han, S. H. Kwon, and B. S. Yoon, (2012), Rapid detection of sacbrood virus in honeybee using ultra-rapid real-time polymerase chain reaction, Journal of Virological Methods, 179(1), p195-200.
[https://doi.org/10.1016/j.jviromet.2011.10.014]

-
Zhang, J., J. Feng, Y. Liang, D. Chen, Z. H. Zhou, Q. Zhang, X. Lu, (2001), Three-dimensional structure of the Chinese Sacbrood bee virus, Science in China, series C, Life Sciences, 44(4), p443-448.
[https://doi.org/10.1007/BF02879612]

Appendix
SUPPLEMENTARY DATA

Chemical synthesized oligo nucleotides containing position for detection of genotyping primer were used for standard DNA construction

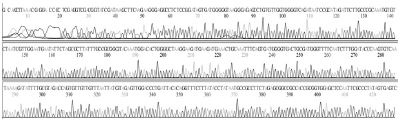
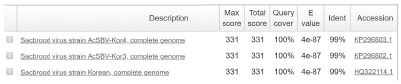
The sequencing result showed SBV genotype 2134D51 was correctly identified from 6 infected honeybee samples
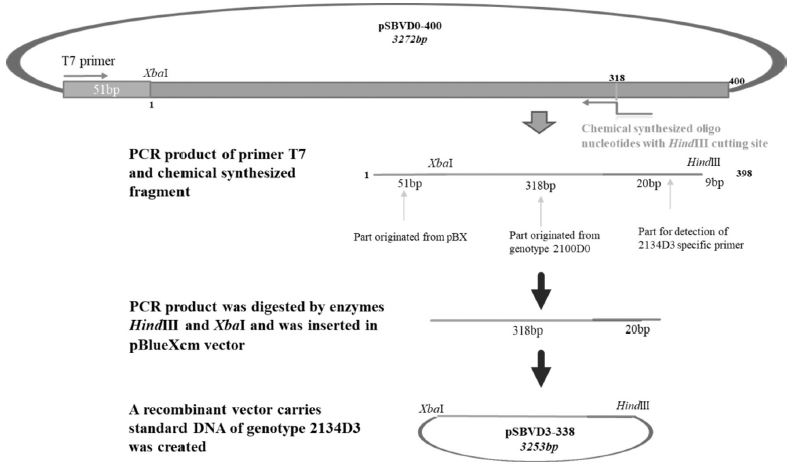
Schematic diagram illustrates the process of standard DNA construction for genotype 2134D3. Recombinant vector pSBVD3-338 carries standard fragment (338 bp) of genotype 2134D3 was constructed from vector pBlueXcm and PCR product that was amplified using chemical synthesized fragment and T7 primer on DNA template from 2100D0 genotype recombinant vector (pSBVD0-400). The restriction enzymes HindIII and XbaI were used.
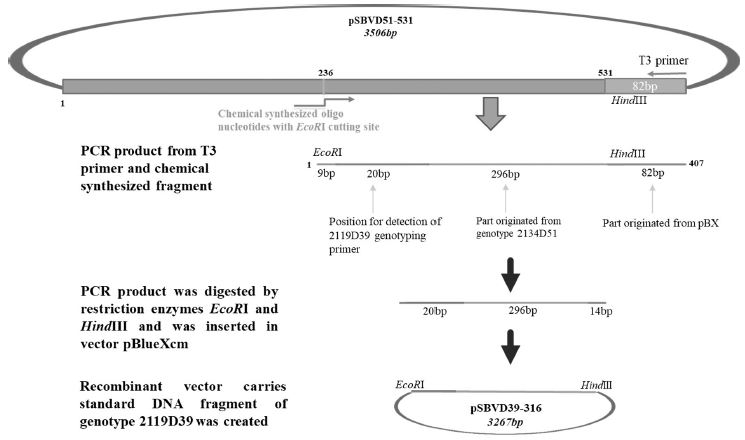
Schematic diagram illustrates the process of standard DNA construction for genotype 2119D39. Recombinant plasmid pBX-SBVD39-316 carries standard fragment (316 bp) of 2119D39 was constructed from vector pBlueXcm and PCR product that was amplified from chemical synthesized fragment and T7 primer on DNA template from 2134D51 genotype recombinant DNA (pSBVD51-531). Restriction enzymes HindIII and EcoRI were used.
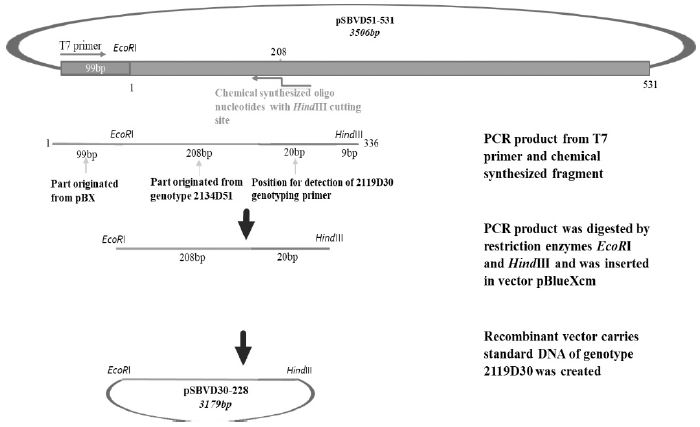
Schematic diagram illustrates the process of standard DNA construction for genotype 2119D30. Recombinant plasmid pSBVD30-228 carries standard fragment (228 bp) of 2119D30 was constructed from vector pBlueXcm and PCR product that was amplified from chemical synthesized fragment and T7 primer on DNA template from 2134D51 standard fragment (pSBVD51-531). Restriction enzymes HindIII and EcoRI were used.