First Report of the Wax Beetle, Platybolium alvearium Blair (Coleoptera: Tenebrionidae) from Nepal
Abstract
Platybolium alvearium is reported for the first time from Nepal. It was found in several localities of the central part of the country. The intraspecific genetic distance of the Cytochrome C oxidase I gene showed 2.7% of genetic distance among samples from Nepal and Vietnam. The phylogenetic relationship between different beetle species associated with honeybees was also discussed.
Keywords:
Platybolium alvearium, darkling beetles, honeybee species, DNA barcoding, NepalINTRODUCTION
Nepal is a country of 147,516 square km, lies between coordinate approximately 28°N and 84°E in the Himalayan region, on the southern flank of the Himalayan mountain range and sharing borders with China in the north and India in the south. Based on the altitude, Nepal can be divided into three main geographical regions: Himalayan region (4000 to 8848 m), Mountain region (500 to 4000 m) and Terai region (75 to 500 m), and majority of honeybee species are found in Terai and Mountain regions of the country (Allen, 1995). Nepal is rich in term of honeybee diversity and distribution of three native open nesting honeybees (Apis florea, Ap. laboriosa and Ap. dorsata), one native close nesting honeybee (Ap. cerana) with different geographical races (Ap. cerana cerana in mountain region and Ap. cerana indica in Terai region) and one exotic honeybee (Ap. mellifera) (Allen, 1995; Devkota et al., 2016; Thapa et al., 2018), made it interesting for studying honeybees biology and pollination.
Colony loss is attributed to the number of biotic and abiotic factors such as decreasing genetic diversity, exposure to agrochemicals and a diminished suitable foraging habitat and pests and diseases (Potts et al., 2010; Goudson et al., 2015; Baude et al., 2016). Recently, an updated list of pests and diseases of honeybees in Nepal was provided by Thapa et al. (2018).
The DNA barcode region is a partial mitochondrial DNA sequence (Cytochrom C oxidase 1 in animals) that is used as a diagnostic market for identification (Hebert et al., 2003). Recently this method has been used for accurate identification of the difficult groups of organisms (Han et al., 2018), cryptic species (Carolan et al., 2012; Shashank et al., 2014; Williams et al., 2015), as well as immature stage of animals.
During recent expedition trip to Central Nepal, we had collected several specimens of darkling beetles associated with Eastern honeybee, Ap. cerana. As there wasn’t any report of beetles in the list of honeybee pests in Nepal, this species was recognized and reported as a new insect species associated with honeybees in Nepal. In addition, the partial sequence of COI gene of this species is deposited in the GenBank to facilitate the identification of this species through DNA barcoding.
MATERIALS AND METHODS
1. Sample collection and morphological identification
The beetles were collected using forceps from several managed hives of Ap. cerana in Kathmandu and Pokhara of the mountain region in Nepal. At the time of sampling, all log hives of Ap. cerana was empty and only managed and wall hives were examined. The colonies were inspected carefully, and the samples were only collected from managed hives and preserved in absolute alcohol and kept in -20 prior to DNA extraction. After DNA isolation, several beetles were pinned and dried for morphological identification using Leica EZ4HD stereomicroscope.
2. DNA extraction and amplification
Total DNA was extracted from the hind leg of each beetle using the DNeasy Blood & Tissue extraction kit (Qiagen, Germany), followed the manufacturer’s protocol. As the LCO-1490 and HCO-2198 primer set (Folmer et al., 1994) failed to amplify the barcoding region; two new primers were developed based on available GenBank sequences of honeybee related beetles (MT602618 for Aethina tumida, MG817071 and MG817069 for Platybolium alvearium, MG817070 and MG817068 for Alphitobius diaperinus) to use for amplification of 369 bp length of barcoding region. The barcoding region of the mitochondrial cytochrome c oxidase I gene was amplified by the Polymerase Chain Reaction (PCR) using AccuPower PCR PreMix (Bioneer, Daejeon, Korea) with the primer set CO-APF (5-GATTCTGACTWCTWCTYCCACCYTC-3) and CO-APR (5-TAWARAATWGGRTATCCTCCYCC-3). PCR reaction conditions included initial denaturation for 5 min at 95℃, 35 cycles of 10 s at 95℃ for denaturation, 30 s at 52℃ for primer annealing, and 30 s at 72℃ for extension, and a final extension for 5 min at 72℃. PCR products were separated on 1% agarose gel, and bands visualized with EcoDye staining (Biofact) and UV transillumination. PCR products were sequenced by Macrogen (South Korea).
3. Sequence analysis
The forward and reverse sequences were edited and assembled using the BIOEDIT v7.0.5.2 (Hall, 1999) to produce a consensus sequence for each sample and all sequences were aligned in MEGA 7 (Kumar et al., 2016) using ClustalW (Thompson et al., 1994). The sequences data of collected beetles from Nepal are publicly available on the GenBank database under accession numbers MT602616 and MT602617.
4. Sequence analysis
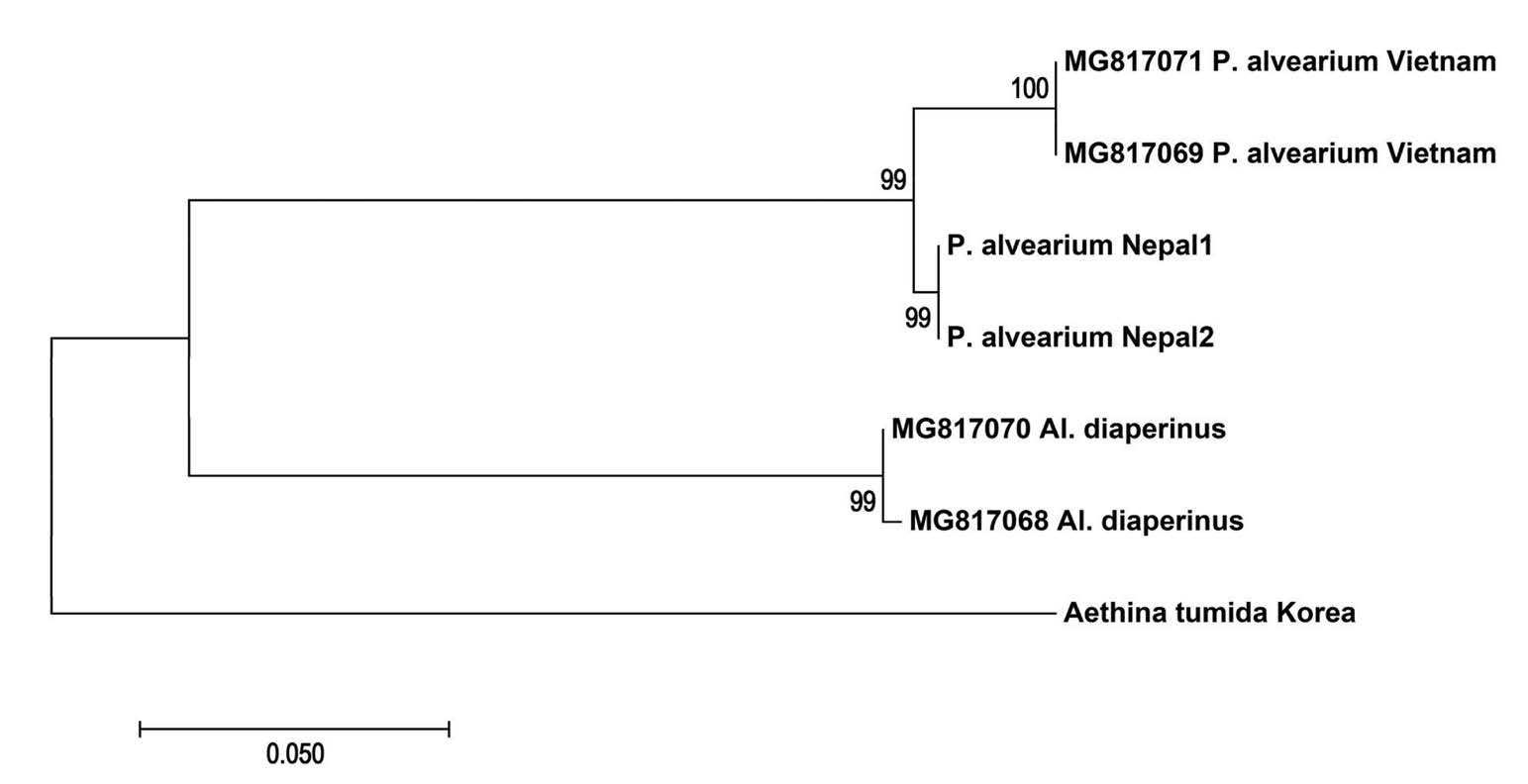
The evolutionary distances between haplotypes were calculated using Kimura’s 2-parameter model (Kimura, 1980) in MEGA7 (Kumar et al., 2016). For phylogenetic analysis, two previously reported sequences of P. alvearium and two sequences of Al. diaperinus were also included. The DNA barcoding part of small hive beetle, Ae. tumida, was also provided for the first time and used to make a phylogenetic tree of the hive related beetles. The jModelTest (version 2.1.3) program was used to select the best nucleotide substitution model using the Default parameters (Darriba et al., 2012). Phylogenetic relationship based on COI gene were conducted using the maximum likelihood (ML) method (Kishino et al., 1990) in Mega7, using GTR+G mutation model and 1000 bootstrap replication was used to evaluate the branching confidence.
RESULTS AND DISCUSSION
1. Identification
Genus Platybolium Blair, 1938Type species, Platybolium alvearium
Platybolium alvearium Blair, 1938 (Fig. 1)

Platybolium alvearium; 1. habitus, dorsal view; 2. habitus, ventral view; 3. Head, enlarged; 4. Beetle on the hive debris.
Diagnosis. Body dark brown; head blackish brown, only anterior two third of clypeus yellowish brown’ antenna 11 segmented with antennal club. Pronotum with lateral margins very narrow, anterior angles only slightly produced; pronotum narrowly yellowish brown laterally; mesocoxal cavity not bordered by a groove posteriorly, legs brown; elytral intervals about equally carinate. Apart from P. alvearium, two more darkling beetles, the black fungus beetle Alphitobius laevigatus and the lesser mealworm Al. diaperinus, are associated with Eastern honeybee (Maitip et al., 2016). P. alvearium can be easily recognizable from these species in having a narrow longitudinal ridge between each elytral stria (Dolson et al., 2019).
Material examined. 2♂, 5♀, 20 km North East Kathmandu, 27.700769°N, 85.300140°E, Jung & Mohamadzade leg., 2020.01.04; 1♂, 6♀, Pokhara, Ghachok village, 28.2096°N, 83.9856°E, Mohamadzade, Thapa & Jung leg., 2020.01.07.
Distribution. Sri Lanka, China, India (Blair, 1938), Bangladesh (Rahana & David, 2019), Vietnam (Dolson et al., 2019), Nepal (New record).
Remarks. P. alvearium is reported for the first time from Nepal. This species was collected from different hives of A. cerana in two localities of central part of the country (Kathmandu and Pokhara). As this species has already been reported from Bangladesh and India, it seems it is widespread throughout the country. In addition, based on the its world distribution, it is expected to find this species in Bhutan, Myanmar, Thailand, Laos and Cambodia (Fig. 2).
This species is only associated with Ap. cerana colonies and there is not any record of finding this species in Ap. mellifera or other wild honeybees’ nest (Pande et al., 2015; Dolson et al., 2019). The biology of this insect is unknown but it supposed to be a scavenger beetle and there is no report of aggressive behavior of Ap. cerana against P. alvearium in Vietnam where the number of beetles inside the colonies was low (Dolson et al., 2019) but in India, Ap. cerana tried to sting beetles in highly infested colonies (Pande et al., 2015). We did not do any statistical analysis on the number of beetles inside each colony, but the number of beetles was low, and no egg, larvae or pupa were found inside the colonies. One of the reasons for this finding is that the hives were examined in January and perhaps winter is not a good season to find immature stages of this beetle.
All sequences of P. alvearium made a well-supported clade (99% of bootstrap value); inside recent group, sequences from Vietnam and Nepal made two well supported subclades (100% bootstrap value) (Fig. 3). Both studied samples of P. alvearium from different localities in Nepal shared same haplotype and the highest intraspecific genetic distance among sequences from Nepal and Vietnam was 2.7%. The similarity rate of the sequences of P. alvearium from Nepal with previously reported sequences from Vietnam was 97.5%. The similarity of the sequences of P. alvearium from Nepal with sequences of Uloma sp. (Accession No. MK075820) from India was 99.4%. There is not any report about association of Uloma with honeybees. On the other hand, the similarity rate of DNA barcoding region of P. alvearium from Nepal with Uloma opacipennis (KJ510014) and U. punctata (KJ003365) is 87.3 and 86.7% respectively. Furthermore, the similarity rate of the Uloma sp. (MK075820) from India with U. opacipennis (KJ510014), U. impressa (HM433276) and Uloma punctata (KJ003365) was 85.3, 85.3, and 85% respectively showing that probably the specimen has been misidentified and this is why we didn’t included this sequence in downstream analysis.
Acknowledgments
This study was supported by the BSRP through the National Research Foundation of Korea (NRF), Ministry of Education (NRF-2018R1A6A1A03024862).
References
-
Allen, M. F. 1995. Bees and beekeeping in Nepal. Bee World 76(4): 185-194.
[https://doi.org/10.1080/0005772X.1995.11099269]

-
Baude, M., W. E. Kunin, N. D. Boatman, S. Conyers, N. Davies, M. A. K. Gillespie, R. D. Morton, S. M. Smart and J. Memmott. 2016. Historical nectar assessment reveals the fall and rise of floral resources in Britain. Nature 530: 85-88.
[https://doi.org/10.1038/nature16532]

- Blair, K. G. 1938. A new genus and species of tenebrionid beetle in bee-hives in India. The Entomologist’s Monthly Mag. 74: 222-223.
-
Carolan, J. C., T. E. Murray, U. Fitzpatrick, J. Crossley, H. Schmidt, B. Cederberg, L. McNally, R. J. Paxton, P. H. Williams and M. J. F. Brown. 2012. Colour patterns do not diagnose species: quantitative evaluation of a DNA barcoded cryptic bumblebee complex. PLoS ONE 7: 662-667.
[https://doi.org/10.1371/journal.pone.0029251]

-
Darriba, D., G. L. Taboada, R. Doallo and D. Posada. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9: 772.
[https://doi.org/10.1038/nmeth.2109]

-
Devkota, K., S. C. Dhakal and R. B. Thapa. 2016. Economics of beekeeping as pollination management practices adopted by farmers in Chitwan district of Nepal. Agric & Food Secure 5(6): 1-6.
[https://doi.org/10.1186/s40066-016-0053-9]

- Folmer, O., M. Black, W. Hoeh, R. Lutz and R. Vrijenhoek. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3: 294-299.
-
Goulson, D., E. Nicholls, C. Botías and E. L. Rotheray. 2015. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science Express 347.
[https://doi.org/10.1126/science.1255957]

- Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95-98.
-
Han, T., S. Kim, H. J. Yoon, I. G. Park and H. Park. 2018. Genetic variations of DNA barcoding region of bumble bees (Hymenoptera: Apidae) from South Korea. Mitochondrial DNA part A 30(1): 30-42.
[https://doi.org/10.1080/24701394.2018.1450396]

-
Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111-120.
[https://doi.org/10.1007/BF01731581]

-
Kishino, H., T. Miyata and M. Hasegawa. 1990. Maximum likelihood inference of protein phylogeny and the origin of chloroplasts. J. Mol. Evol. 31(2): 151-160.
[https://doi.org/10.1007/BF02109483]

-
Kumar, S., G. Stecher and K. Tamura. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33: 1870-1874.
[https://doi.org/10.1093/molbev/msw054]

-
Maitip, J., Z. Zhang, K. Tan, P. H. Thai, M. V. Nabozhenko, A. G. Kirejtshuk, P. Chantawannakul and P. Neumann. 2016. A scientific note on the association of black fungus beetle (Alphitobius laevigatus, Coleoptera: Tenebrionidae) with Eastern honey bee colonies (Apis cerana). Apidologie 48: 271-273.
[https://doi.org/10.1007/s13592-016-0471-5]

-
Pande, R., N. S. A. Thakur, S. V. Ngachan and D. J. Rajkhowa. 2015. First record of wax beetle, Platybolium alvearium Blair (Coleoptera: Tenebrionidae), in Eastern Himalaya: A new threat to Indian honey bee (Apis cerana Fabricius) colonies. J. Ent. Res. 39(3): 269-273.
[https://doi.org/10.5958/0974-4576.2015.00033.X]

-
Potts, S. G., J. Biesmeijer, C. Kremen, P. Neumann, O. Schweiger and W. E. Kunin. 2010. Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol. 25: 345-353.
[https://doi.org/10.1016/j.tree.2010.01.007]

- Rahana, P. A. and C. V. David. 2019. Study on pests and predators of Apis cerana indica F. in selected apiaries of Thrissur district, 31st Kerala Science Congress, 2-3 February, Kollam, 200.
-
Shashank, P. R., A. K. Chakravarthy, B. R. Raju and K. R. M. Bhanu. 2014. DNA barcoding reveals the occurrence of cryptic species in host-associated population of Conogethes punctiferalis (Lepidoptera: Crambidae). Appl. Entomol. Zool. 49: 283-295.
[https://doi.org/10.1007/s13355-014-0248-0]

-
Thapa, R., S. Aryal and C. Jung. 2018. Beekeeping and honey hunting in Nepal: current status and future perspectives. pp. 111-127. in Asian beekeeping in the 21st centry, eds. by P. Chantawannakul, G. Williams and P. Neumann, Springer, Singapore.
[https://doi.org/10.1007/978-981-10-8222-1_5]

-
Thompson, J. D., D. G. Higgins and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680.
[https://doi.org/10.1093/nar/22.22.4673]

-
Williams, P. H., A. M. Byvaltsev, B. Cederberg, M. V. Berezin, F. Odegaard, C. Rasmussen, L. L. Richardson, J. X. Huang, C. S. Sheffield and S. T. Williams. 2015. Genes suggest ancestral colour polymorphisms are shared across morphologically cryptic species in arctic bumblebees. PLoS ONE 10(12): e0144544.
[https://doi.org/10.1371/journal.pone.0144544]

Appendix
DNA barcoding region of the COI gene of Platybolium alvearium from Nepal
P. alvearium voucher CONE01 (accession no. MT602616)
CTTCCACCTTCATTAACACTTCTGCTAATAAGAAGAATTGTTGAAAGAGGAGCGGGTACAGGATGAACAGTGTACCCTCCACTTTCATCCAATATCGCACACGGAGGATCCTCCGTTGATTTAGCAATTTTTAGATTACATTTAGCAGGAATTTCTTCCATCCTAGGAGCCGTAAACTTCATTACTACAGTAATTAATATACGTCCTCAAGGAATATCATTTGATCGAATACCTTTATTTGTATGAGCAGTAGTAATTACTGCTGTTCTTCTTCTTCTTTCTCTTCCCGTACTAGCCGGAGCAATCACTATACTCTTAACAGACCGAAATATTAATACATCCTTCTTTGACCCTGCAGGAGGAGGAGAC
P. alvearium voucher CONE02 (accession no. MT602617)
CTTCCACCTTCATTAACACTTCTGCTAATAAGAAGAATTGTTGAAAGAGGAGCGGGTACAGGATGAACAGTGTACCCTCCACTTTCATCCAATATCGCACACGGAGGATCCTCCGTTGATTTAGCAATTTTTAGATTACATTTAGCAGGAATTTCTTCCATCCTAGGAGCCGTAAACTTCATTACTACAGTAATTAATATACGTCCTCAAGGAATATCATTTGATCGAATACCTTTATTTGTATGAGCAGTAGTAATTACTGCTGTTCTTCTTCTTCTTTCTCTTCCCGTACTAGCCGGAGCAATCACTATACTCTTAACAGACCGAAATATTAATACATCCTTCTTTGACCCTGCAGGAGGAGGAGAC
Aethina tumida (accession no. MT602618)
AACTTTATATTTCATTTTTGGTATTTGATCAGGCATAGTAGGAACTTCATTAAGACTCCTAATTCGAACTGAATTAGGAAATCCTGGGTCATTAATTGGAAATGACCAAATTTACAATGTTATTGTTACAGCTCACGCTTTCATTATAATTTTCTTTATAGTTATACCATTTATAATTGGTGGATTCGGAAACTGATTAGTTCCATTAATATTAGGAGCCCCTGATATAGCTTTCCCTCGAATGAATAATATAAGATTCTGACTTTTACCACCATCCCTTTCTCTTCTACTTATAAGAAGAATTGTAGAAAGAGGAGCAGGAACAGGATGAACAGTGTACCCTCCACTTTCATCTAATATCGCTCATGGTGGATCTTCAGTTGATTTAGCTATTTTTAGACTTCACTTAGCAGGTATTTCTTCTATTTTAGGTGCAGTAAATTTTATTACTACTGTAATTAATATGCGACCCTCAGGCATAACCTTTGATCGAATACCTTTATTTGTTTGAGCTGTAGTAATTACAGCTATCCTTCTTCTACTTTCATTACCTGTATTAGCAGGAGCTATTACTATACTACTAACAGATCGAAATCTAAATACTACTTTCTTCGACCCATCGGGAGGGGGTGATCCAATCCTATACCAACACTTATTT