
Survey and Molecular Detection of Melisococcus plutonius, the Causative Agent of European Foulbrood in Honeybee Apis mellifera in the Jammu Region of India
Abstract
The study was carried out during 2016-17 in different locations of Jammu and Kashmir, India. Infections caused by the European foulbrood disease organism Melisococcus plutonius were recorded from a minimum of 6% (Pranu) to a maximum of 16.66% (Sartingal ) in colonies of the Doda district and from a minimum of 1.47% (Parnote) to a maximum of 15.18% (Digdol) in the district of Ramban. Incidences of European foulbrood were recorded maximally in September (14.59%) and minimally in March (1.81%). Percentages of European foulbrood infections of experimental colonies of the university apiary were lowest (1.81%) in March and maximal (15.37%) in September. A polymerase chain reaction (PCR) molecular diagnosis for EFB was conducted on 60 larval samples collected from different areas of the four districts for PCR amplification of partial 16S rRNA gene fragments (486 bp). Results showed that out of 60 only 6 samples (10%) were positive for M. plutonius in different apiaries. The study documented the occurrence of EFB by PCR assay in apiaries of different regions of Jammu and Kashmir for the first time.
Keywords:
European foulbrood, Molecular characterization, Apis melliferaINTRODUCTION
Honeybee colony losses are an increasing problem throughout the world (Biesmeijer et al., 2006; Johnson, 2007; Potts et al., 2010a, b; Neumann and Carreck, 2010; Vanbergen, 2013; Powney et al., 2019). Managed honey bees are the most important commercial pollinators and account for 35% of the global food production (William and Anderson, 1974; Free, 1993; Roubik, 1995; Delaplane and Mayer, 2000, Morandin and Winston, 2006; Klatt et al., 2014). The honeybee diseases usually persist as unapparent infections and cause no overt signs of disease. They can dramatically affect honey bee health and shorten the lives of infected bees under certain conditions (Bailey et al., 1983; Hansen and Brødsgaard, 1999; Forsgren et al., 2009; Okamura, 2016). The pests and pathogens are the single most important cause of otherwise inexplicable colony losses leading to Colony Collapse Disorder (Paxton, 2010; Dainat et al., 2012; Jeroen et al., 2013; Bojana et al., 2014).
The state of Jammu & Kashmir is blessed with all the four species of honeybees viz., Apis florea F., A. dorsata F., A. cerana F., A. mellifera L. and several geographical races/ecotypes are expected to exist (Abrol, 2012). The state offers great potential for both migratory and non-migratory beekeeping (Abrol, 2012). Over the past 20 years, honey bees have been faced with many threats including pests and diseases which has led to the listing of honey bee diseases in OIE (world organization for animal health) Terrestrial Animal Health Code. Honeybees are subject to attack by many viruses (Bailey and Ball, 1991; Allen and Ball, 1996), five of which are associated with Varroa and one with tracheal mites. There are two main bacterial diseases found in Asian honeybees: American Foul Brood disease (AFB) caused by Paenibacillus larvae (Oldroyd and Wongsiri, 2006) and European Foul Brood (EFB) caused by Melissococcus plutonius (Bailey and Collins, 1982). In India, European foulbrood disease appeared for the first time in A. cerana during 1970 and then disappeared after killing 25~30 percent of the colonies in the Karad area of Maharashtra (Diwan et al., 1971; Bailey, 1974). However, in A. mellifera the disease was noticed in Himachal Pradesh and the Dharwad area of Karnataka during 1998 killing 25~40 percent of the colonies in some apiaries. 1998 (Anonymous, 1998; Viraktamath, 1998). Then in A. cerana, the disease was again noticed in 2002 after an absence of three decades killing about 60 percent of the colonies in Himachal Pradesh. Death of the brood was also recorded at prepupal and pupal stages (Rana et al., 2004).
European foulbrood disease prevalence in A. mellifera in northern India is posing a serious threat to the bee keeping industry by affecting colony productivity, colony multiplication and antibiotic residue in honey. There is every chance of this disease spreading to other parts of the country where A. cerana and A. mellifera exist, and affecting the local beekeeping industry. Some studies have already been done in India on this disease in A. cerana (Rana et al., 2004) and its occurrence, symptomatology, cultural, epidemiology and chemical control in A. mellifera (Bahman and Rana, 2002).
It is not possible to identify the disease exactly based on symptomology as most of the brood diseases have symptoms more or less similar. Several methods have been used to detect M. pluton. These include microscopic examination of carbol fuchsin stained smears of diseased larvae (Hornitzky and Wilson, 1989), culture of bacteria from infected larvae or honey (Hornitzky and Smith, 1998), scanning electron microscopy (Alippi, 1991) and enzyme-linked immunosorbent assays (Pinnock and Featherstone, 1984). However, for isolation of M. pluton, specific and sensitive detection methods are required to ensure its absence from bee products for export purposes and for epidemiological studies. Evidently, molecular techniques can help to reliably detect, identify and characterize honeybee pathogens so that a diagnosis of the disease is initiated well in time to save the beekeeping industry before a catastrophe occurs. The prevalence of these and other new pathogens and their potential correlation with bee losses in Jammu & Kashmir at present is unknown. No information is available on molecular characterization of Melissococcus plutonius of A. mellifera in the Jammu region. A hemi-nested polymerase chain reaction (PCR) developed by Djordjevic et al. (1998) for the detection of Melissococcus plutonius in adult bees and honey bee products could be used for the molecular characterization. Keeping this in view, the study was carried out with the objectives to survey the seasonal incidence of European Foul Brood disease in the Jammu region and its molecular characterization in A. mellifera L.
MATERIALS AND METHODS
The present investigations were conducted during August 2016 to April 2017 in A. mellifera colonies maintained at the experimental field of Division of Entomology and in apiaries of private beekeepers in several regions of Jammu division (Fig. 1).
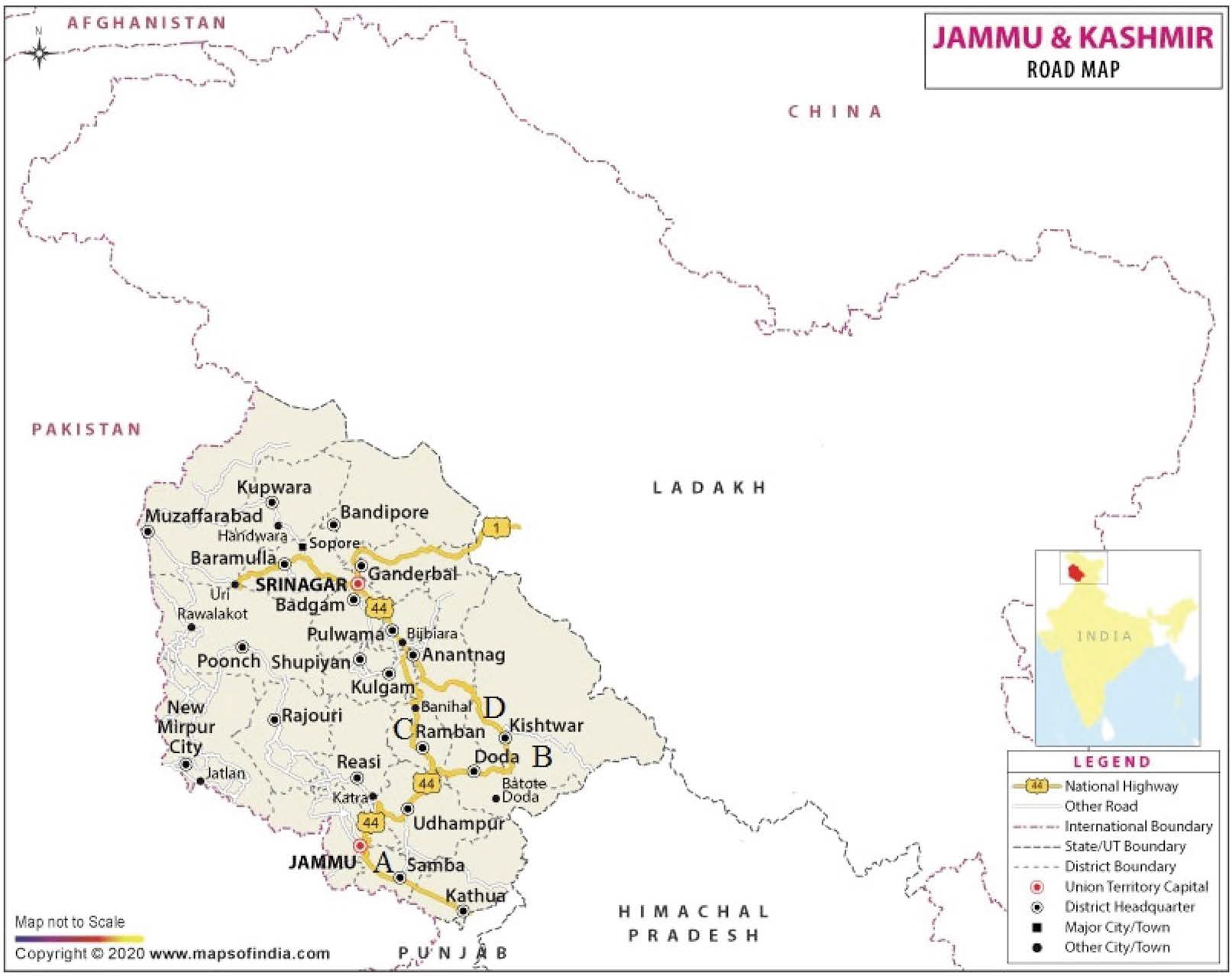
Map showing EFB inspection areas in Jammu region. EFB has been detected in all the districts. A=Jammu, B=Doda, C=Ramban, D=Banihal.
1. Incidence of European foulbrood disease
European foulbrood disease was recorded in A. mellifera during August 2016 to April 2017 in the university apiary and in apiaries of private beekeepers. Severity of European foulbrood disease was studied with regard to the percent colony infection in the apiary and percent brood infection by using the following formulae:
The strength of each colony was estimated by observing the number of combs covered with bees of respective species.
Brood area was measured with the help of measuring grid having squares, each square measuring 3 cm2. The brood area was converted into cm2 by multiplying the number of squares with a factor of 3.
2. Symptomology
A. mellifera colonies exhibiting typical symptoms of European foulbrood disease were observed during the period of experimentation. The symptoms were recorded on alternate day with respect to stage, change in colour, position, shape, odour and texture of the brood in the diseased colonies at University Apiary. Five colonies of A. mellifera were selected from each apiary in the regions of Jammu, Ramban, Banihal and Doda districts which were expected to have occurrence of diseases. The disease incidence was recorded at an interval of 30 days.
3. Samples
Five 4~5 day old larval samples showing clinical signs of EFB were collected from each apiary for the molecular characterization of the bacterium M. plutonius causing EFB. Bee larvae were collected using sterile forceps and the larvae were put in 70% ethanol for further studies.
4. Molecular detection
Molecular level detection of the present isolates was done through polymerase chain reaction (PCR) technique as it is a sensitive, rapid and economical based technique for detecting the disease, European foulbrood microorganism which requires no antiserum as well no monoclonal antibodies. The molecular studies were initiated by the isolation of total DNA from the diseased samples and further confirmed by PCR based gene specific primers.
5. Methods to extract DNA from bee larvae
Sixty samples of 4~5 day old larvae showing clinical signs of EFB were collected from apiaries from different areas of the Jammu region, five samples from each apiary using sterile forceps and then the larvae were put in 70% ethanol for the further process of molecular characterization of the bacterium M. plutonius causing EFB.
(1) Isolation of total DNA
The total DNA extracted from collected larvae by CTAB (cetyl trimethylammonium bromide) based method (Protocol for the isolation of DNA) were prepared from a pool of 5 larvae of A. mellifera by adopting the following method:
Extraction buffer was prepared by 2% hexadecyl trimethyl ammonium bromide (CTAB), (1.4 M NaCl, 1% polyvinylpyrrolidone, 0.02 M ethylenediaminetetraacetic acid, 0.1 M Tris-HCl, and incubated for 5 min at 65℃ in a dry block heater. 750 μL of the extraction buffer was put into each Eppendorf tube, and then the individual samples were macerated in a sterile pestle mortar into paste and then added to the Eppendorf tubes containing extraction buffer. 20 μL of mercaptaethanol was added in each Eppendorf tube and mix it thoroughly. Then the Eppendorf tubes were incubated for 10 min in a hot air oven at 65℃. After incubation, 500 μL C : I (Chloroform : isopopanol) in the ratio of 24 : 1 is added into each Eppendorf tube and mixed thoroughly. The Eppendorf tubes are then centrifuged at 12000 rpm for 15 min. The transparent liquid was then transferred into fresh Eppendorf tubes and the resultant supernatant was then discarded. The nucleic acids were precipitated with 100 μL of sodium acetate (3 M, pH 5.2) and 500 μL of ice-cold ethanol (100%). The suspension was incubated at -20℃ for 20 min and then centrifuged at 15000 rpm for 20 min. The resultant supernatant was discarded from each Eppendorf tube and the pellet was washed twice with 50 μL ethanol (70%), air dried and resuspended in 50 μL of TE buffer (10 mM TrisHCl, pH 8.0, 1 mM EDTA) in each Eppendorf tube. DNA samples were then stored at -20℃.
(2) Checking of DNA
Presence of DNA was checked in 0.8 percent agarose gel, prepared in 1×TBE (Tris borate EDTA) buffer and stained with ethidium bromide (0.4 μL). DNA (3 μL) was loaded with DNA loading dye (bromophenol) and electrophoresed at 115 V for 30~40 minutes and visualized in UV trans illuminator.
(3) Purification of DNA
The DNA visualized in UV trans illuminator had presence of a large amount of RNA. So for the purification of DNA from RNA, the samples were treated with RNAse (10 mg/mL). 1 μL of RNAse was taken and added to the target sample. Then 500 μL of autoclaved water was added to the target sample and mixed thoroughly. The sample is incubated at 37℃ for 1 hour. 500 μL of P : C : I (25 : 24 : 1) was added and mixed thoroughly. The samples were centrifuged at 12000 rpm for 15 minutes. Only the upper layer was taken and transferred to the fresh tubes and the lower layer was discarded. 500 μL of C : I (24 : 1) was added to the fresh Eppendorf tubes. The eppendoef tubes are then centrifuged at 12000 rpm for 15 min. The transparent liquid was then transferred into fresh Eppendorf tubes and the resultant supernatant was then discarded. The nucleic acids were precipitated with 100 μL of sodium acetate (3 M, pH 5.2) and 500 μL of ice-cold ethanol (100%). The suspension was incubated at -20℃ overnight and then centrifuged at 15000 rpm for 20 min. The resultant supernatant was discarded from each Eppendorf tube and the pellet was washed twice with 50 μL ethanol (70%), air dried and resuspended in 50 μL of TE buffer (10 mM TrisHCl, pH 8.0, 1 mM EDTA). DNA samples were then stored at -20℃ and 3 μL was used in the PCR.
(4) Detection of M. plutonius by polymerase chain reaction (PCR)
For detection of M. plutonius, PCR was used to amplify a portion of the M. plutonius 16S rRNA gene which comprised of primer MP1, 5′ CTTTGAACGCCTTAGAGA 3′; and primer MP2, 5′ ATCATCTGTCCCACCTTA 3′ (Djordejevic et al., 1998).
Polymerase chain reaction was used to amplify DNA performed in a 25 μL reaction volume containing 5 ng template DNA, 0.2 mM of each dNTP, 0.2 μM of each primer, 1.5 mM MgCl2, 1X PCR buffer and 1 unit Taq polymerase. The reactions were carried out under following temperature and time as given below: Initial Denaturation (95℃) for 2 min, Denaturation (95℃) for 30 sec, Annealing (61℃) for 15 sec, Elongation (72℃) for 60 sec and Extension (72℃) for 5 min.
The PCR amplification was performed in Gene AMP PCR system 9700 Thermal cycler (Applied Bio-Systems, USA). Further, the amplicons so generated were viewed under UV trans lluminator and photographed in Flour-S Multiimager.
6. Statistical analysis
Various statistical tools used in the experiment were mean, standard deviation and 95% confidence interval (Sokal and Rohlf, 2013).
RESULTS
The results of investigation are presented under following headings as under.
1. Symptomology
During the period of investigations, following typical symptoms of the diseases were recorded.
- i) In A. mellifera, diseased bee colonies, colour of the infected larvae (brood) was pale yellow while healthy brood appeared glistening shiny white.
- ii) The healthy brood was in coiled position at the bottom of comb cells while 20% of the infected larvae (pale yellow coloured) were found slightly displaced in upward or downward direction from their actual position.
- iii) In general, death of brood was recorded at pupal stage (>70). About 15 percent occurred at larval stages and rarely in prepupal stage (about 5%), which resulted perforation in sealed brood. However, the colour of the brood did not change which exhibited typical symptoms like those of Thai sac brood virus disease and mite infestation.
- iv) The infected larvae were slightly pulpy or watery. In the advanced stage of the infection, the emitted sour or vinegar like smell.
- v) Remains of the dried brood were noticed in the form of soft scales.
2. Disease incidence
The observations recorded on the incidence of European foulbrood A. mellifera in areas of Jammu, Ramban, Banihal and Doda districts of Jammu region are given in Table 1. The colony infection of European foulbrood disease caused by M. plutonius was recorded from a minimum of 6% (Pranu) to a maximum of 16.66% (Sartingal ) in colonies of district Doda.
The present investigations revealed that the percent brood infection of European foulbrood disease in A. mellifera in areas of Jammu, Ramban, Banihal and Doda districts of Jammu region are given in Table 2. Brood infection percentages caused by European foulbrood ranged from a minimum of 1.48% (Parnote) to a maximum of 15.25% (Digdol) of district Ramban.
Incidence of European foulbrood in university apiary from July, 2016 to April, 2017 was found to be maximum in September (14.59%) and minimum in March (1.88%) (Table 3).
Percent brood infection of European foulbrood in university apiary was recorded from a minimum of 1.82% (March) to a maximum of 15.38% (September), as given in Table 4.
The bacteria present in the larval samples which caused disease in A. mellifera was detected and confirmed through polymerase chain reaction assay. A polymerase chain reaction (PCR) molecular diagnosis for EFB was conducted on 60 larval samples collected from different areas of the four districts for PCR amplification of partial 16S rRNA gene fragments (486 bp). Two pairs of M. plutonius specific primers (MP1& MP2 and MP1 & MP3 each reported by Djordejevic et al., 1998), were used to amplify the different regions of the present M. plutonius genomes. Out of the two primer pairs, 1 primer pair (MP1 & MP3) reported by Djordojevic et al. (1998) amplified M. plutonius genome of A. mellifera. The amplicons (amplified product) were observed as clear distinct bands under UV transilluminator. These primers were found specific as resulted in the amplification of genome with amplicons of expected size (486 bp) (Figs. 2 and 3).
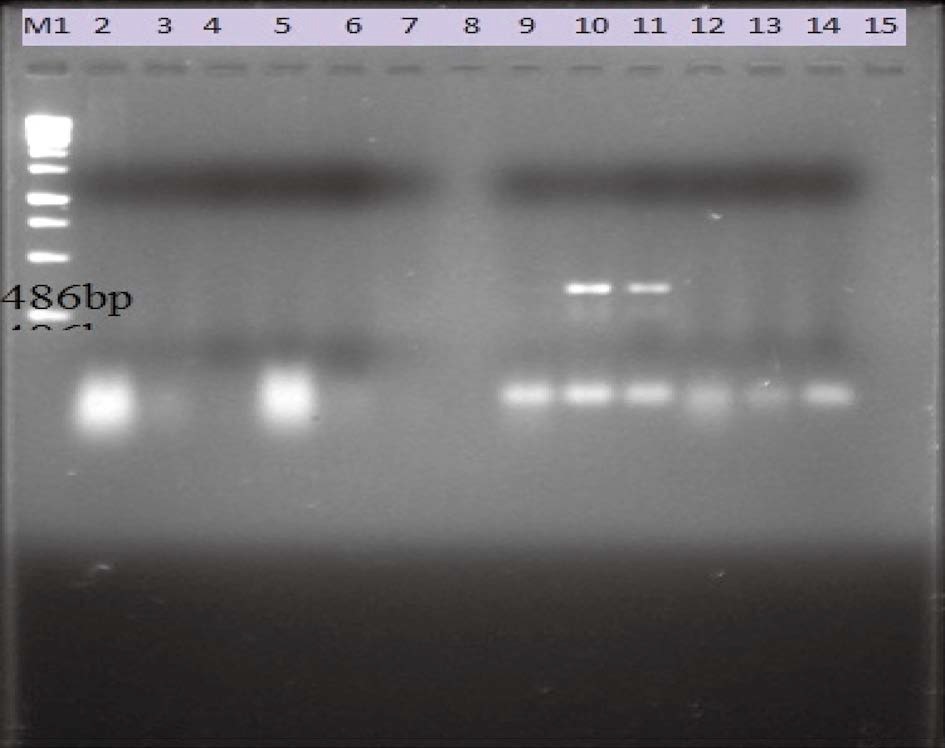
Agarose gel electrophoresis (1.5%) of products of the first generation 16S rRNA PCR using primers MP1 and MP2. The DNA fragment was amplified from DNA extractions of larvae with European foulbrood. Lane 1, 250 bp molecular markers. Lane 2, 3, 4, 5, 6 - pranu colonies, Lane 7, 8, 9, 10, 11 - sartingal colonies, Lane 12, 13, 14, 15 - attalgarh colonies, Lane 10, 11 - 2 ng M. plutonius DNA.
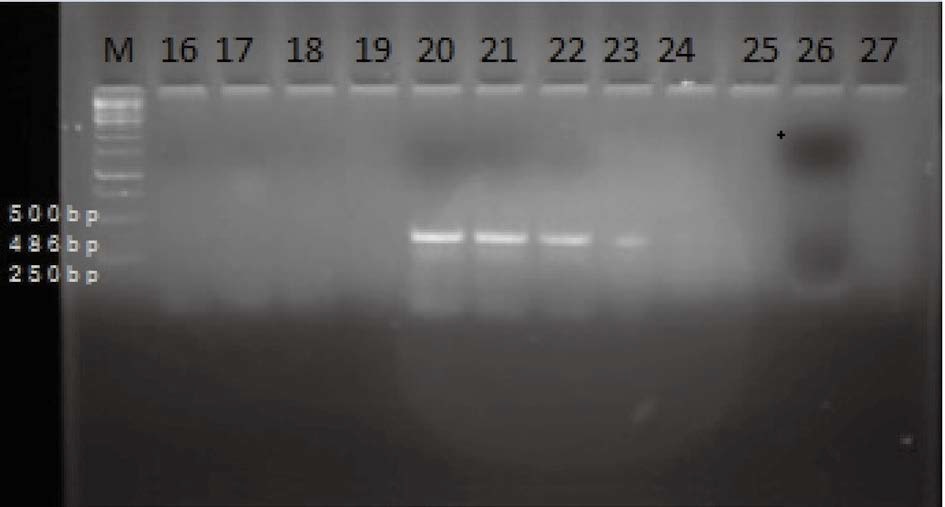
Agarose gel electrophoresis (1.5%) of products of the first generation 16S rRNA PCR using primers MP1 and MP3. The DNA fragment was amplified from DNA extractions of larvae with European foulbrood. Lane 1, 250 bp molecular markers. Lane 16, 17 - Dalwah colonies, Lane 18, 19 - Parnote colonies, Lane 20, 21 - Digdol colonies, Lane 22, 24 - miran sahib Lane 23, 25 - Arbal Lane 26, 27 - Shataninallah, Lane 20, 21, 22, 23 - 2 ng M. plutonius DNA.
Results showed that out of 60 only 6 samples (10%) were positive for M. plutonius in different apiaries. This study document the occurrence of EFB by PCR assay in apiaries of different regions of Jammu (Table 5).
DISCUSSION
The discussion of investigation are presented under suitable headings as under.
1. Symptomatology
The present study conducted on the symptoms of European foulbrood disease in A. mellifera found that in A. mellifera diseased bee colonies, colour of the infected larvae (brood) was pale yellow while healthy brood appeared glistening shiny white. The healthy brood was in coiled position at the bottom of comb cells while some of the infected larvae (pale yellow coloured) were found slightly displaced in upward or downward direction from their actual position. About 70~80 percent death of brood was recorded at pupal stage. Sometimes about 15 percent occurred at larval stages and rarely in pre-pupal stage (about 5%), which resulted perforation in sealed brood. However, the colour of the brood did not change which exhibited typical symptoms like those of Thai sac brood virus disease and mite infestation. Also the infected larvae were slightly pulpy or watery. It was also revealed that in the advanced stage of the infection, their emitted sour or vinegar like smell. Remains of the dried brood were noticed in the form of soft scales. Similar results were reported by Forsgen (2013) who conducted study on field diagnosis of EFB based on the visual inspection of brood combs and detection of diseased larvae. He observed that in a colony suffering from EFB there was irregular capping of the brood capped and uncapped cells being found scattered irregularly over the brood frame (known as pepper pot brood). The youngest larvae that die from the infection cover the bottom of the cell and are almost transparent, with visible trachea. Older larvae die mal positioned and flaccid in their cells; twisted around the walls or stretched out lengthways. The colour of affected larvae changes from pearly white to pale yellow, often accompanied by a loss in segmentation. More advanced symptoms can manifest as further colour changes to brown and greyish black, sometimes ultimately leaving a dark scale that is more malleable than those typically found with American foul breed.
2. Disease Occurrence
In the present study, it was observed that the colony infection of European foulbrood disease caused by Melisococcus plutonius was recorded from a minimum of 6% (Pranu) to a maximum of 16.66% (Sartingal ) in colonies of district Doda. The study got support from Mwale (1992) who examined 343 hived Apis mellifera colonies in 36 apiaries at 19 locations in Malawi and found that 8% of the colonies were infected by Melissococcus plutonius.
It was evident from the findings of the study that brood infection of European foulbrood was recorded from a minimum of 1.48% (Parnote) to a maximum of 15.25% (Digdol) of district Ramban. Similar study was conducted by Rao et al. (2011) who studied on characterization of Melissococcus plutonius from European honey bee (Apis mellifera) of North-West Himalayas and found that Melissococcus plutonius, the causal bacterium of European foulbrood (EFB) disease was isolated from A. mellifera L. of North-West Himalayas. The disease affected 2.52 to 18.52% brood in 2.65 to 15.00% honey bee colonies.
3. Seasonal incidence of European foulbrood disease in university apiary
Based on the observation and results of the study, incidence of European foulbrood in university apiary from July, 2016 to April, 2017 was found to be maximum in September (14.59%) and minimum in March (1.88%) and the percent brood infection of European foulbrood in university apiary was recorded from a minimum of 1.82% (March) to a maximum of 15.38% (September). These results got support from the findings of Rao (2009) who conducted study on molecular characterization of Melissococcus plutonius of hive honey bees and its control with antibiotics and revealed that maximum incidence of disease was recorded in September 2007 (18.52%) and declined completely by March.
4. Molecular detection through Hemi-nested polymerase chain reaction (PCR) technique
Based on the observation and results of the study, the bacteria present in the larval samples which caused disease in A. mellifera was detected and confirmed through polymerase chain reaction assay. A polymerase chain reaction (PCR) molecular diagnosis for EFB was conducted on 60 larval samples collected from different areas of the four districts for PCR amplification of partial 16S rRNA gene fragments (486 bp). Two pairs of M. pluton specific primers (MP1 & MP2) and (MP1 & MP3) each reported by Djordejevic et al. (1998), were used to amplify the different regions of the present M. pluton genomes. Out of the two primer pairs, 1 primer pair (MP1 & MP3) reported by Djordjevic et al. (1998) amplified M. pluton genome of A. mellifera. The amplicons (amplified product) were observed as clear distinct bands under UV transilluminator. These primers were found specific as resulted in the amplification of genome with amplicons of expected size (486 bp). Results showed that of the 60 samples only, 6 samples (10%) were positive for M. plutonius in different apiaries. This study documents the occurrence of EFB by PCR assay in apiaries of different regions of Jammu. Similar study was conducted by Djordjevic et al. (1998) who conducted study on development of a hemi-nested PCR assay for the specific detection of Melissococcus plutonius and in his study a pair of oligonucleotide primers (MP1 & MP2) was used for the polymerase chain reaction (PCR) amplification of a 486 base pair (bp) fragment of the 16S rRNA gene of 26 geographically diverse Australian M. plutonius (causative agent of European foulbrood) isolates. PCR primers spanning a region of the 16S rRNA gene from position 893~1377 failed to amplify a product when template DNA from a wide range of pathogenic and saprophytic bacteria were used including Paenibacillus larvae, Enterococcus faecium and Spiroplasma melliferum. The PCR did, however reliably amplify a 486 bp fragment (when the annealing temperature was lowered by 5℃) using template DNA isolated from the phylogenetically-related bacterium Enterococcus faecalis. PCR amplicons generated from E. faecalis and M. pluton were readily distinguished by digestion with the restriction endonuclease Hinfl and electrophoresis in 1.5% agarose or by electrophoresis in 1% agarose containing bisbenzidene/polyethylene glycol. A hemi-nested PCR requiring a combination of primers MP1 and a third primer, MP3, which spanned 25 nucleotides from position 1168~1144 and internal to the 486 bp amplicon generated by primers MP1 and MP2 was developed. The hemi-nested PCR amplified a 276 bp M. plutonius specific product that was not amplified with E. faecalis DNA. In sensitivity studies, the PCR assay could reliably detect approximately 1~10 organisms/ mi. This level of sensitivity was achieved using crude DNA templates (boiled cell lysate) prepared using lnsta-gene matrix. The PCR assay could also detect M. plutonius in brood with European foulbrood. These findings got support from Khezri et al. (2016) who studied on the prevalence of M. plutonius by polymerase chain reaction in Kurdistan apiaries, Iran and in his study for multiplex PCR amplification of partial 16S rRNA (=SSU rRNA) gene fragments (486 bp), was conducted on 100 samples. Results showed that of 100 samples only 4 samples (4%) were positive for M. plutonius in Kurdistan apiaries.
The colony infection of European foulbrood disease caused by Mellisococcus plutonius was recorded from a minimum of 6% (Pranu) to a maximum of 16.66% (Sartingal) in colonies of Doda region. Percent brood infection of European foulbrood was also recorded from a minimum of 1.47% (Parnote) to a maximum of 15.18% (Digdol) of Ramban region.
Incidence of European foulbrood in university apiary from July, 2016 to April, 2017 was found to be maximum in September (14.59%) and minimum in March (1.81%). Similarly incidence of sac brood was found maximum in March (9.09%) and minimum in August (2.07%). Percent brood infection of European foulbrood in university apiary was recorded from a minimum of 1.81% (March) to a maximum of 15.37% (September). A polymerase chain reaction (PCR) molecular diagnosis for EFB was conducted on 60 larval samples collected from different areas of the four regions for PCR amplification of partial 16S rRNA gene fragments (486 bp). Results showed that of the 60 samples, only 6 samples (10%) were positive for M. plutonius in different apiaries. This study document the occurrence of EFB by PCR assay in apiaries of different regions of Jammu. It is thus concluded that surveys are required to characterize the distribution and prevalence of M. plutonius in different areas of Jammu district.
Acknowledgments
The authors thank Dr. Susheel Sharma, Assistant Professor School of Biotechnology for his help during the course of investigation. We also thank anonymous reviewers for their valuable suggestions on earlier draft of this paper.
References
- Abrol, D. P. 2009. Bees and beekeeping in India. 2nd Edn Kalyani Publishers, Ludhiana India.
- Abrol, D. P. 2012. Pollination biology: Biodiversity conservation and agricultural production. Springer Dordrecht Heidelberg London, New York.
-
Alippi, A. M. 1991. A comparison of laboratory techniques for the detection of significant bacteria of the honey bee, Apis mellifera, in Argentina. J. Apic. Res. 30: 75-80.
[https://doi.org/10.1080/00218839.1991.11101237]

-
Allen, M. and B. Ball. 1996. The incidence and world distribution of honey bee viruses. Bee World 77: 141-162.
[https://doi.org/10.1080/0005772X.1996.11099306]

- Anonymous. 1998. Studies on control of natural enemies and diseases of Honey bees. Annual Progress Report, Department of Entomology and Apiculture, Dr. Y.S. Parmar University of Horticulture and Forestry, Nauni, Solan, 45-46.
- Bahman, S. and B. S. Rana. 2002. Incidence of the foulbrood disease in Apis mellifera L. colonies at Solan, Himachal Pradesh. Pest Manag. & Econ. Zool. 10(1): 87-91.
-
Bailey, L. 1974. An unusual type of Streptococcus pluton from the Eastern hive bee. J. Invert. Pathol. 23: 246-247.
[https://doi.org/10.1016/0022-2011(74)90192-X]

-
Bailey, L. and B. Ball. 1991. Honey Bee Pathology. Academic Press, London.
[https://doi.org/10.1016/B978-0-12-073481-8.50006-0]

-
Bailey, L., B. V. Ball and J. N. Perry. 1983. Association of viruses with two protozoal pathogens of the honey bee. Ann. Appl. Biol. 103: 13-20.
[https://doi.org/10.1111/j.1744-7348.1983.tb02735.x]

-
Biesmeijer, J. C., S. P. M. Roberts, M. Reemer, R. Oholemuller, M. Edwards, T. Peeters, A. P. Schaffers, S. G. Potts, R. Kleukers, C. D. Thomas, J. Settele and W. E. Kunin. 2006. Parallel declines in pollinators and insect-pollinated plants in Britain and the Netherlands. Science 313: 351-354.
[https://doi.org/10.1126/science.1127863]

- Bojana, B., J. Marko and S. Jonel. 2014. Honey bee colony collapse disorder (Apis mellifera L.) - possible causes. Scientific Papers Series Management, Econ. Engin. Agric. & Rural Dev. 14(2): 13-18.
-
Dainat, B., D. Van Engelsdorp and P. Neumann. 2012. Colony Collapse Disorder in Europe. Environ. Microbiol. Rep. 4(1): 123-125.
[https://doi.org/10.1111/j.1758-2229.2011.00312.x]

-
Delaplane, K. S. and D. F. Mayer. 2000. Crop Pollination by Bees. CABI Publishing, New York.
[https://doi.org/10.1079/9780851994482.0000]

- Diwan, V. V., K. K. Kshirsagar, A. V. Ramanrao, D. Raghunath, C. S. Bhambure and S. H. Godbole. 1971. Occurrence of new bacterial disease of Indian honey bee Apis indica F. Curr. Sci. 40: 196-297.
-
Djordjevic, P., S. L. Noone and M. Hornitzky. 1998. Development of a hemi nested PCR assay for the detection of Melissococcus pluton. J. Apicult. Res. 37(3): 165-174.
[https://doi.org/10.1080/00218839.1998.11100968]

-
Forsgren, E., J. R. de Miranda and M. Isaksson. 2009. Deformed wing virus associated with Tropilaelaps mercedesae infesting European honey bees (Apis mellifera). Exp. Appl. Acarol. 47: 87-97.
[https://doi.org/10.1007/s10493-008-9204-4]

- Free, J. B. 1993. Insect pollination of crops. Academic Press, London-New York.
-
Hansen, H. and C. J. Brødsgaard. 1999. American foulbrood: a review of its biology, diagnosis and control. Bee World 80: 5-23.
[https://doi.org/10.1080/0005772X.1999.11099415]

-
Hornitzky, M. A. Z. and L. Smith. 1998. Procedures for the culture of Melissococcus pluton from diseased brood and bulk honey samples. J. Apic. Res. 37: 293-294.
[https://doi.org/10.1080/00218839.1998.11100987]

-
Hornitzky, M. A. Z. and S. Wilson. 1989. A system for the diagnosis of the major bacterial brood diseases. J. Apic. Res. 28: 191-195.
[https://doi.org/10.1080/00218839.1989.11101183]

- Johnson, R. 2007. Recent Honey Bee Declines. Congressional Research Service, Washington, DC: 14 pages.
-
Klatt, B. K., A. Holzschuh, C. Westphal, Y. Clough, I. Smit, E. Pawelzik and T. Tscharntke. 2014. Bee pollination improves crop quality, shelf life and commercial value. Proc. R. Soc. B 281: 20132440.
[https://doi.org/10.1098/rspb.2013.2440]

-
Morandin, L. A. and M. L. Winston. 2006. Pollinators provide economic incentive to preserve natural land in agroecosystems. Agric. Ecosyst. Environ. 116: 289-292.
[https://doi.org/10.1016/j.agee.2006.02.012]

-
Neumann, P. and C. Carreck. 2010. Honey bee colony losses: a global perspective. J. Apic. Res. 49: 1-6.
[https://doi.org/10.3896/IBRA.1.49.1.01]

-
Okamura, B. 2016. Hidden Infections and Changing Environments. Integ. Comp. Biol. 56(4): 620-629.
[https://doi.org/10.1093/icb/icw008]

- Oldroyd, B. P. and S. Wongsiri. 2006. Asian Honey Bees Biology, Conservation and Human Interactions. Harvard University Press, Cambridge.
-
Paxton, R. J. 2010. Does infection by Nosema ceranae cause “Collony Colapse Disorder” in honey bees (Apis mellifera)?. J. Apic. Res. 49(1): 80-84.
[https://doi.org/10.3896/IBRA.1.49.1.11]

-
Pinnock, D. E. and N.E. Featherstone. 1984. Detection and quantification of Melissococcus pluton infection in honeybee colonies by means of enzyme-linked immunosorbent assay. J. Apic. Res. 23: 168-170.
[https://doi.org/10.1080/00218839.1984.11100627]

-
Potts, S. G., S. P. M. Roberts, D. Robin, G. Marris, M. A. Brown, R. Jones, P. Neumann and J. Settele. 2010. Declines of managed honey bees and beekeepers in Europe. J. Apic. Res. 49(1): 15-22.
[https://doi.org/10.3896/IBRA.1.49.1.02]

-
Potts, S. G., J. C. Biesmeijer, C. Kremen, P. Neumann, O. Schweiger and W. E. Kunin. 2010a. Global pollinator declines: trends, impacts and drivers. Trends Ecol. Evol. 25: 345-353.
[https://doi.org/10.1016/j.tree.2010.01.007]

-
Powney, G. D., C. Carvell, M. Edwards, K. A. Roger, R. K. A. Morris, H. E. Roy, B. A. Woodcock and N. B. J. Isaac. 2019. Wide spred losses of pollinating insects in Britain. Nat. Comm. 10, Article number: 1018.
[https://doi.org/10.1038/s41467-019-08974-9]

- Rana, B. S., R. Rana, G. Kumar, H. K. Sharma, J. K. Gupta and K. Dayal. 2004. European foulbrood causing havoc to the native bee, Apis cerana F. in Himachal Pradesh. Pest Manag. & Econ. Zool. 12(1): 109-111.
- Rao, K. M. 2009. Molecular characterization of Melissococcus plutonius of hive honey bees and its control with antibiotics, Ph.D. Thesis. Department of Entomology and Apiculture, Dr. YS Parmar University of Horticulture and Forestry, Solan. 97.
- Rao, K. M., B. S. Rana, S. K. Chakravarty, S. Katna and A. Sharma. 2011. Characterization of Melissococcus pluton from European Honey bee (Apis mellifera L.) of North-West Himalayas. Intl. J. Sci. & Nat. 2: 632-638.
- Roubik, D. W. 1995. Pollination of cultivated plants in the tropics. FAO Agricultural Services Bulletin 118, Rome.
- Sokal, R. R. and F. J. Rohlf. 2013. Biometry: The principles and practices of statistics in biological research 4th Edition. W. H. Freeman and Company Ltd., San Francisco, USA.
-
Vanbergen, A. J. 2013. Threats to an ecosystem service: pressures on pollinators. Frontiers Ecol. Environ. 11(5): 251-259.
[https://doi.org/10.1890/120126]

-
van der Sluijs, J. P., N. Simon - Delso, D. Goulson, L. Maxim, J. M. Bonmatin and L. P. Belzunces. 2013. Neonicotinoids, bee disorders and the sustainability of pollinator services. Curr. Opinion Environ. Sustain. 5(3-4): 293-305.
[https://doi.org/10.1016/j.cosust.2013.05.007]

- Viraktamath, S. 1998. A bacterial disease of Apis mellifera L. in Dharwad. Insect Env. 4: 27.
- William, P. N. and L. Anderson. 1974. Insect pollinators frequenting strawberry blossoms and the effect of honey bees on yield and fruit quality. J. Amer. Soc. Hort. Sci. 99(1): 40-44.
