
Over-expression and Purification of RNA Dependent RNA Polymerase from Black queen cell virus in Honeybee
Honeybee populations infected by numerous viruses could not easily distinguish from the healthy populations because there are no apparent clinical symptoms and signs of infection. Black queen cell virus (BQCV) is one of the most prevalent viruses, resulting in the death of queen larvae and pupae. Like to other picornaviruses, BQCV has the RNA dependent RNA polymerase (RdRp) that catalyzes the replication of viral RNA. The optimal conditions for expression of MBP-BQCV-RdRp protein were confirmed by an induction with 0.1mM IPTG at 25°C for 6 hours and the MBP-BQCV-RdRp protein was purified using by amylose resin. Total yield of protein reached approximately 5mg per 50ml of induced cells. Due to its property that is an essential enzyme of virus replication, purified recombinant BQCV-RdRp proteins can be provided to the further lead as an important source to study how to control the BQCV viral disease in honeybee, development of a rapid kit for the diagnosis of BQCV infection in field,or finding out the anti-viral chemical compound for the inhibitor assay to prevent BQCV replication in honeybee.
Keywords:
Black queen cell virus (BQCV), RNA dependent RNA polymerase (RdRp), Maltose binding protein (MBP), HoneybeeINTRODUCTION
Honeybee plays a vital role in the development of beekeeping by providing valuable products including beeswax, propolis, pollen and royal jelly. In addition, many agricultural crops require them for pollination to increase the crop yield. However, the occurrence of several honeybee diseases infected by pathogens including viruses and bacteria is considered as a threat to the beekeeping industry. Black queen cell virus (BQCV) is identified as one member of the “picorna-like” group of viruses and belongs to the new genus Cripavirus (Family Dicistroviridae) (MayO, 2002). It was originally detected in dead honeybee queen larvae and pupae (Bailey and Wood, 1977). In Australia, BQCV has been reported to be the most common cause of death of queen larvae (Anderson, 1993).
The complete genome of BQCV is a single-stranded RNA of 8550 nucleotides including two Open reading frames (ORFs) with the specificity of each regions, the 5’ proximal ORF encodes a putative replication protein, while the 3’ proximal ORF encodes a capsid polyprotein (Leat et al., 2000).
In the replication of the viral genome, the RNA dependent RNA polymerase (RdRp) plays an important role in ensemble with host proteins and viral other proteins (Buck, 1996; Lai, 1998). Previous studies on comparison of structures and the phylogeny of the viral RNA dependent RNA polymerases confirmed the identification of a new conserved motif apparently present in all RdRps (Bruenn, 2003; Koonin, 1991). RdRp was validated as a suitable taxonomic marker for the assignment of members of the order Picornavirales, with the potential for use independently of other genetic or phenotypic marker (Baker and Declan, 2008). In Korea, phylogenetic analysis of Black queen cell virus genotypes showed that BQCV may infect both A. cerana and A. mellifera (Noh et al., 2012).
This study aimed to express and purify RNA dependent RNA polymerases from Black queen cell virus in E.coli system. This purified RdRp protein will be used as a potential source of antigen for generating monoclonal antibodies to develop a commercial rapid kit which can detect rapidly and sensitively BQCV in honeybee.
MATERIALS AND METHODS
Honeybee sample and total RNA extraction
Honeybee samples infected by BQCV were collected in 2013 in South Korea. Collected honeybee samples were homogenized by MagNA Lyser green Beads (Roche, Switzerland) then were extracted total RNA by using AllspinTM for total DNA & RNA isolation from tissues and cultured cells (GeneAll-Korea), according to the manufacturer’s instructions. Finally, RNAs were eluted in 50ul of sterilized RNAse-free water and were stored at -70°C until used. Concentration of total RNAs were determined by Biophotometer machine (Eppendorf-Germany).
Reverse transcription
The total extracted RNAs were reverse transcribed by AccuPower® RT Premix (Bioneer-Korea). RT reaction was performed with 2ug total RNA and 200 pmole of OligodT (Bionics-Korea) for 42°C, 60 mins and 94°C, 5 mins. Subsequently, cDNAs can be used for experiments.
Molecular cloning of RNA dependent RNA polymerase (RdRp) gene from BQCV
The primer set for BQCV-RdRp was designed base on the published nucleotide sequences (GenBank Accession No. EF517515.1). Details of primers are shown in Table 1. The amplified DNA fragment was cloned into pGEM-3Zf(+) vector (Promega-USA). BQCV-RdRp clones were verified by DNA-sequencing. Afterward, the BQCV-RdRp were subcloned into pMAL c2 expression vector (New England Biolabs). The recombinant pMal-BQCV-RdRp were examined by restriction analysis on 1.5% agarose gel, then were transformed to E.coli BL21 (DE3) pLysS for expression.
Expression of MBP-BQCV-RdRp protein
MBP-BQCV-RdRp in E.coli BL21 (DE3) pLysS were cultured in LB medium including Ampicillin (0.05mg/ml) and Chloramphenicol (0.05mg/ml) at 37°C for overnight and then 1ml of bacteria culture was immediately subcultured into 25ml LB medium. To optimize conditions for protein expression, bacterial cultures were grown with different optical densities at 600nm (OD600) and induced with different concentrations of IPTG. In addition, the different temperatures for IPTG induction were examined at 25°C and 37°C. After 1, 3, 6, 12 and 24 hours, aliquots of induced bacterial cells were collected by centrifugation and the cells were suspended in 1X PBS buffer then disrupted by sonication. The expression of MBP-BQCV-RdRp was evaluated by SDS-PAGE.
Purification of recombinant MBP-BQCV-RdRp protein by amylose resin
MBP-BQCV-RdRp were purified using amylose bind agarose resin (Elpis-Korea) that was designed for rapid purification of recombinant protein fused with MBP. The MBP-BQCV-RdRp binded to amylose, which was immobilized on the agarose resin before. After unbound proteins were washed away, the MBP-BQCV-RdRp were recovered by elution with maltose containing elution buffer. Concentration of the purified MBP-BQCV-RdRp was determined by Bradford assay with BSA standard (1mg/ml) and was stored at -70°C until used.
RESULTS AND DISCUSSION
Amplification of BQCV-RdRp fragment
BQCV-RdRp gene was amplified by PCR reaction with specific primer pairs (Table 1). Amplified PCR fragment was cloned into pGEM-3Zf(+) vector and the clone was named as pGEM-BQCV-RdRp. BQCV-RdRp clone was screened by BamHI/SalI restriction analysis (Fig. 1). The sequencing result revealed 93% homology of nucleotide sequences (Fig. 2) and 98% homology of amino acid sequences to deposited sequences (Genbank accession No. EF517515.1) (Fig. 3).
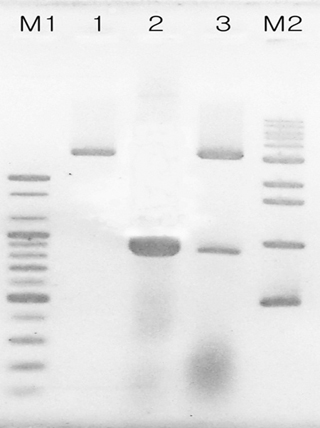
Confirmation of pGEM-BQCV-RdRp clone by restriction analysis. Lane M1 is 100 bp DNA ladder (Bioneer-Korea: 100, 200, 300, 400, 500, 600, 700, 800, 900, 1000, 1200, 1600, 2000 bp). Lane M2 is 1kb DNA ladder (Bioneer- Korea: 500, 1000, 1600, 2000, 2961, 4000, 5007, 5991, 8000, 10200 bp). Lane 1 is pGEM-3Zf(+) vector which was digested by BamHI/SalI. Lane 2 is PCR product PCR product amplified with BQCV-RdRp-F-BamHI and BQCV-RdRp-R-SalI primer set. recombinant pGEMBQCV-RdRp DNA which was digested by BamHI/SalI. It is clear to observe BQCV-Rdrp that was digested from clone with an expected size around 903 bp and pGEM vector with an expected size around 3179 bp in lane 3. This result is compared with pGEM vector and BQCV-Rdrp which were showed in lane 1 and lane 2.

Nucleotide sequence homology between Genbank EF517515.1 and BQCV Korean strain (pGEM-BQCV-RdRp). The sequence determined was corresponding to RNA dependent RNA polymerase region of EF517515.1 deposited in NCBI databases and symbolized by asterisk (*) in the consensus line. Alignment to EF517515.1 revealing 93% homology.
Expression of MBP-BQCV-RdRp protein
BQCV-RdRp was subcloned into pMAL-C2 vector, then transformed to E.coli BL21 (DE3) pLysS host cell. Based on amino acid sequence, the size of MBP-BQCVRdRp was calculated around 80 kDal. Firstly, the expression of MBP-BQCV-RdRp protein was evaluated both in pellet and supernatant under a normal condition at 25°C induced by 0.6mM IPTG during 6hrs. The result confirmed that MBP-BQCV-RdRp protein expressed well both in pellet and supernatant with expected bands around 80 kDal (Fig. 4). In this experiment, pQE30-gfp-ELP was used as a positive control and expression of pMAL-C2 was performed as a negative control with expected bands about 50 kDa.
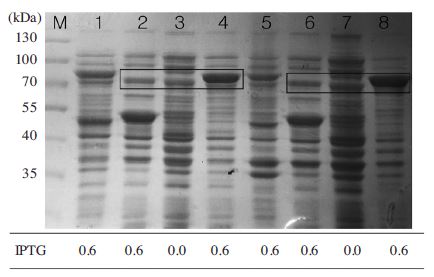
Expression of MBP-BQCV-RdRp protein in E.coli BL21 (DE3) pLySs. After sonication, location of MBP-BQCVRdRp protein was examined both in the supernatant and pellet. Lane M is protein size marker (Thermo scientific-USA:170, 130, 100, 70, 55, 40, 35, 25, 15 and 10 kDal). Lane 1 and 5 are induced pQE30-gfp-ELP from supernatan and pellet, respectively. Lane 2 and 6 are pMal-C2 induced IPTG from supernatant and pellet, respectively. Lane 3 and 7 are MBP-BQCV-RdRp protein without IPTG from supernatant and pellet, respectively. Lane 4 and 8 are induced MBP-BQCV-RdRp protein from supernatant and pellet, respectively. The expected bands around 80 kDal of MBP-BQCV-RdRp protein were marked in two boxes and compared with negative controls in lanes 2, 3 and lanes 6, 7.
Optimization expression conditions of MBPBQCV-RdRp protein
Firstly, the optical densities of the cell were examined (Fig. 5). To establish the optimum conditions of subculture, MBP-BQCV-RdRp were grown with different optical density at 600nm (OD600) values then induced the cells with 0.6mM IPTG at 25°C for 6 hours. In new study, an induction at OD600 = 0.8 was determined as an optimized condition for production of MBP-BQCV-RdRp protein.
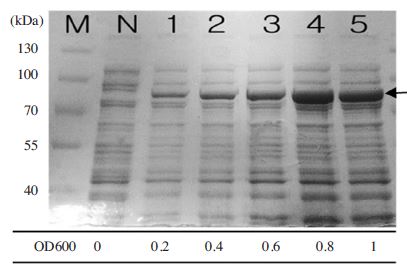
Optimal OD600 value for expresion of MBP-BQCV-RdRp protein. MBP-BQCV-RdRp proteins in E.coli BL21 (DE3) pLysS were started to induce with 0.6mM IPTG at variable OD600 concentrations. Inducted cells were collected after 6 hours. Lane M is protein size marker (Thermo scientific-USA; 170, 130, 100, 70, 55, 40, 35, 25, 15 and 10 kDal). Lane N is a negative control as no induction. Lane 1 to 5 are induced protein at OD600 = 0.2; 0.4; 0.6; 0.8 and 1, respectively. The black arrow implied the expected protein bands. MBP-BQCV-RdRp protein expressed highest at OD600 = 0.8.
Under an optimal OD600 value, IPTG concentration and induced time were evaluated. The results pointed out 0.1mM IPTG was enough to express well MBP-BQCVRdRp protein, whereas a higher IPTG concentration seemed not to increase dramatically expression levels (Fig. 6). Afterward, incubation time for induction was confirmed that 6 hours was optimal (Fig. 7) and the recombinant BQCV-RdRp could be degraded after long incubations. It is clear to observe that MBP-BQCV-RdRp protein expressed highly at 0.1mM IPTG for 6 hours induction.
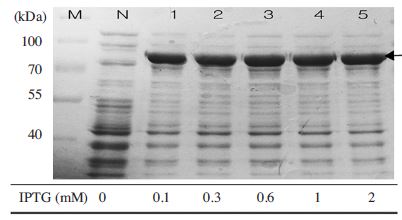
Expresion of MBP-BQCV-RdRp protein depends on IPTG concentration. MBP-BQCV-RdRp in E.coli BL21 (DE3) pLysS was cultured until OD600 = 0.8 and induced by variable IPTG concentrations. Induced cells were collected after 6 hours. Lane M is protein size marker (Thermo scientific-USA; 170, 130, 100, 70, 55, 40, 35, 25, 15 and 10 kDal). Lane N is a negative control as no induction. Lane 1 to 5 are induction at 0.1, 0.3, 0.6, 1 and 2mM IPTG, respectively. The black arrow implied the expected protein bands. This result confirmed that 0.1mM IPTG was enough for expression.
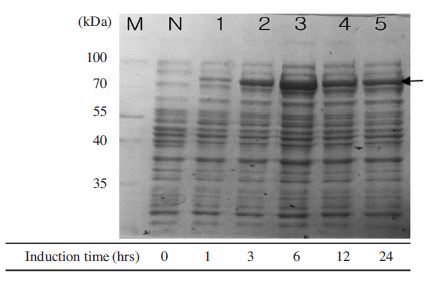
Optimal induction time for expression of MBP-BQCVRdRp protein. The expression of recombinant protein was performed by 10% SDS-PAGE and 1X TGS (Tris/Glycine /SDS) running buffer. Proteins were heated denaturation before loading on the gel. Induction of MBP-BQCV-RdRp protein in E.coli BL21 (DE3) pLysS with 0.1mM IPTG was started at OD600 = 0.8 and cells were collected after different induction time. Lane M is protein size marker (Thermo scientific-USA; 170, 130, 100, 70, 55, 40, 35, 25, 15 and 10 kDal). Lane N is a negative control without induction. Lane 1 to 5 are induced protein at 1, 3, 6, 12 and 24 hours, respectively. The black arrow implied expected protein bands. This result confirmed that 6 hours was an optimal condition for induction time.
Initially, IPTG induction was performed at 25°C. Whether expression of MBP-BQCV-RdRp protein is depend on temperature, MBP-BQCV-RdRp protein was induced with two different temperatures. As the result, MBP-BQCV-RdRp protein expressed at 25°C better than 37°C (Fig. 8). Compare with other researches, RdRp gene could be expressed well at 30°C (Zhou et al., 2006; Sankar and Porter, 1991). On sum up, the optimal conditions for expression of MBP-BQCV-RdRp protein were pointed out as OD600 = 0.8 with 0.1mM IPTG for 6 hours at 25°C induction.
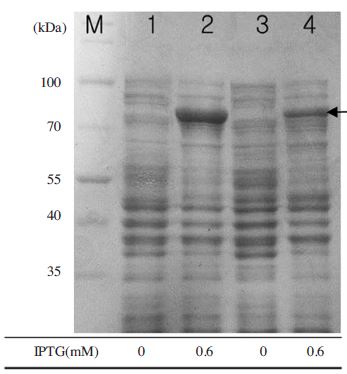
Optimal induction temperature for expression of MBPBQCV- RdRp protein. Under optimal conditions of OD600, IPTG concentration, time for induction, the evaluation of MBP-BQCV-RdRp protein in E.coli BL21 (DE3) pLysS was performed at 25°C and 37°C. Lane M is protein size marker (Thermo scientific-USA; 170, 130, 100, 70, 55, 40, 35, 25, 15 and 10 kDal). Lane 1 and 3 are negative controls as no induction at 25°C and 37°C, respectively. Lane 2 is MBP-BQCV-RdRp protein induced at 25°C and lane 4 is MBP-BQCV-RdRp protein induced at 37°C. The black arrow implied the expected protein bands. In this case, induction temperature at 25°C was better than 37°C for expression of MBP-BQCV-RdRp protein.
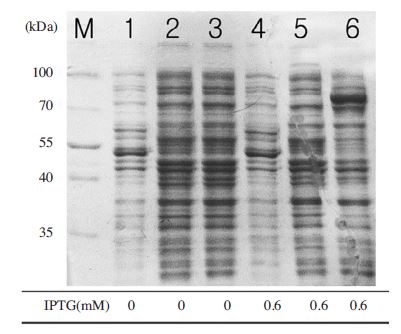
Comparison of expression levels of BQCV-RdRp in expression vector systems. The expression of recombinant proteins were performed by 10% SDS-PAGE and 1X TGS (Tris/Glycine/SDS) running buffer. All samples were loaded after sonication and heated denaturation. Lane M is protein size marker (Thermo scientific-USA; 170, 130, 100, 70, 55, 40, 35, 25, 15 and 10 kDal). Lane 1,2,3 are negative controls of pQE30-BQCV- RdRp-ELP, pET32 a+-BQCVRdRp and MBP-BQCV-Rdrp without induction, respectively. Lane 4, 5, 6 are BQCV-RdRp-ELP, pET32 a+- BQCV-RdRp and MBP-BQCV-RdRp induced at 25°C, respectively. The black arrow implied the expected protein bands. The expected band was observed clearly in lane 6 about 80 kDal. There were no expressed bands of BQCVRdRp protein in pQE30-BQCV-RdRp-ELP and pET32 a+- BQCV-RdRp.
Comparison of expression levels of BQCV-RdRp in expression vector systems
BQCV-RdRp gene were subcloned to pQE30-gfp-ELP, pET 32 a+ and pMAL-C2 expression vector systems then examination of expression levels of protein were performed under a minimum condition. Expected protein bands were observed on SDS-PAGE of pQE30-BQCV-RdRp-ELP, pET 32 a+-BQCV-RdRp and MBP-BQCV-RdRp are 94,7 kDal; 32 kDal; 80 kDal, respectively. Subcultured cells were induced at 25°C with 0.6 mM IPTG for 6 hrs. The results demonstrated that BQCV-RdRp could be expressed well in pMal C2 vector (Fig. 9). However, there were no any expected bands of BQCV-RdRp that showed on pQE30-gfp-ELP and pET 32 a+ systems.
Purification of MBP-BQCV-RdRp protein
The pMal C2 vector expressed the target protein with MBP-tag at N-terminal as a fusion protein. Base on this system, the recombinant protein could be purified using amylose resin. As shown in fig. 10, the MBP-BQCVRdRp protein was recovered at a purified form. After purification, amylose resin will be taken out and saved at 4°C for next purification times. Protein concentrations were checked by Bradford assay using BSA (1mg/ml) as a standard. A purified MBP-BQCV-RdRp protein yield was calculated approximately 5mg per 50ml of inducted cells. This yield seem to be a good yield when compare to previously reported results for recombinant protein yield (Nguyen et al., 2009; Nguyen et al., 2010).
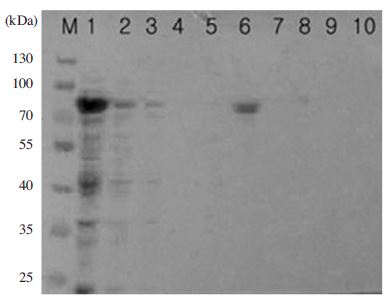
Purification of MBP-BQCV-RdRp protein using amylose binding resin. Protein after purification was determined by 10% SDS-PAGE and 1X TGS (Tris/Glycine/SDS) running buffer. 16ul of protein was loaded on gel after sonication and heated denaturation. Lane M is protein size marker (Thermo scientific-USA; 170, 130, 100, 70, 55, 40, 35, 25, 15 and 10 kDal). Lane 1 is inducted total protein. Lane 2 is a part of protein unbound to amylose resin. Lane 3 is washing by column buffer. Lane 4 is washing by column buffer in 1% Tween 20. Lane 5 is washing by column buffer. Lane 6 is eluted MBP-BQCVRdRp protein. Lane 7 is washing by distilled water. Lane 8 is washing by 0.1% SDS. Lane 9 is washing by distilled water. Lane 10 is washing by column buffer. After purification, one clear band located in lane 6 as a purified MBP-BQCV-RdRp protein form without any other proteins from the cell, this band showed the expected size of MBP-BQCV-RdRp protein about 80 kDal.
Black queen cell virus was originally found in dead honeybee queen larvae and pupae and was known to be the most common cause of death of queen larvae. RdRp is an essential protein required for replication of RNA from initial RNA. Studies of RdRp contribute significantly to understand the replication mechanism of RNA viruses and extend new assays against virus. In this study, the RNA dependent RNA polymerase of Black queen cell virus was successfully amplified. From sequencing results, BQCVRdRp clone revealed 93% homology of nucleotide sequences and 98% homology of amino acid sequences to deposited sequences (Genbank accession No. EF517515.1). RdRp was well expressed in E.coli system and clearly purified by amylose binding resin. The recombinant BQCV-RdRp was highly expressed under optimal conditions at OD600 = 0.8 with 0.1mM IPTG for 6 hours at 25°C induction. It could be used as an antigen to generate monoclonal antibodies for rapid detection BQCV. Morever, RdRp will be used in drug assay to find out chemicals that inhibit the activity of this gene.
Acknowledgments
This work was supported by Bio-Industry Technology Development Program (312027-3) and Advanced Production Technology Development Program (112042-03) and Kyonggi University’s Graduate Research Assis-tantship 2013.
References
- Anderson, D. L., (1993), Pathogens and queen bees, Australian Beekeeper, 94, p292-296.
-
Baker, C.A., and C.S. Declan, (2008), The use of RNA-dependent RNA polymerase for the taxonomic assignment of Picorna - like viruses (order Picornavirales) infecting Apis mellifera L. Populations, J. Virol.
[https://doi.org/10.1186/1743-422X-5-10]

-
Bailey, L., and R.D. Wood, (1977), Two more small RNA viruses from honey bees and further observations on sacbrood and acute bee-paralysis viruses, J. Gen. Virol, 37, p175-182.
[https://doi.org/10.1099/0022-1317-37-1-175]

-
Bruenn, A.J., (2003), A structural and primary sequence comparison of the viral RNA-dependent RNA polymerases, Nucleic Acids Research, 7, p1821-1829.
[https://doi.org/10.1093/nar/gkg277]

- Buck, K.W., (1996), Comparison of the replication of positive-stranded RNA viruses of plants and animals, Adv. Virus Res, 47, p159-251.
-
Koonin, E.V., (1991), The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses, J. Gen. Virol, 72, p2197-2206.
[https://doi.org/10.1099/0022-1317-72-9-2197]

- Lai, M.M., (1998), Cellular factors in the transcription and replication of viral RNA genomes: A parallel to DNA-dependent RNA transcription, J. Virol, 244, p1-12.
- Leat, N., B. Ball, V. Govan, and S. Davison, (2000), Analysis of the complete genome sequence of black queen cell virus, a picorna-like virus of honey bees, J. GenVirol, 81, p2111-2119.
- MayO, M.A., (2002), Virus taxonomy-Houston 2002, Arch Virol, 147, p1071-1076.
- Nguyen, T.K.C., M.S. Yoo, C.H. Yun, and B.S. Yoon, (2009), Over-expression of Major Royal Jelly protein 1 from Apis mellifera L. in E.coli, Korean J. Apiculture, 24(3), p167-173.
- Nguyen, V.P., J.N. No, M.S. Yoo, Y.H. Park, and B.S. Yoon, (2010), Over-expression of Major Royal Jelly protein 3 from Apis mellifera in E.coli, Korean J. Apiculture, 25(3), p163-171.
-
Noh, H.J., K.E. Reddy, S.E. Choe, M.S. Yoo, H.T.T. Doan, C.H. Kweon, M. Ramya, B.S. Yoon, L.T.K. Nguyen, T.T.D. Nguyen, D.V. Quyen, S.C. Jung, K.Y. Chang, S.W. Kang, (2012), Phylogenetic analysis of black queen cell virus genotypes in South Korea, J. Virus Genes.
[https://doi.org/10.1007/s11262-012-0859-x]

- Sankar, S., and A.G. Porter, (1991), Expression, purification, and properties of recombinant Encephalomyocarditis virus RNA-dependent RNA polymerase, J. Virol, 6, p2993-3000.
-
Tentcheva, D., L. Gauthier, N. Zappulla, B. Dainat, F. Cousserans, M.E. Colin, M. Bergoin, (2004), Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite population in France, Appl. Environ Microbiol, 7, p7185-7191.
[https://doi.org/10.1128/AEM.70.12.7185-7191.2004]

-
Zhou, L., L. Jiamin, W. Xiaochun, J. Hong, Y. Fuming, H. Yuanyang, (2006), Expression and characterization of RNA-dependent RNA polymerase of Dendrolimus punctatus Tetravirus, J. Biol. Chem, 5, p571-577.
[https://doi.org/10.5483/BMBRep.2006.39.5.571]


