Evaluation of Quantitative Real-Time Pcr Reference Genes for the Investigation of Gene Expression Profiles in Honeybee Developmental Stages
Abstract
In addition to its role as an essential pollinator, the Western honeybee, Apis mellifera, is regarded as a good model insect due to its highly evolved sociality, labor division, and flexibility of colony management. Therefore, to identify the molecular mechanisms involved in various colony compositions and flexibility of worker division, it is essential to investigate the gene expression in various developmental stages of the honeybee. For target gene quantification, quantitative real-time PCR (qRT-PCR) has been widely used. However, a reliable reference gene stably expressing across different samples should be selected. Thus, to identify credible reference genes in honeybee colonies, we evaluated five candidate genes (RPS5, RPS18, GAPDH, ARF1, and RAD1a) in samples collected from seven developmental stages (egg, 1st instar larvae, 3rd instar larvae, 5th instar larvae, pupa, nurse, and forager) using three analysis programs algorithms (geNorm, NormFinder, and BestKeeper). Although the reference gene ranked slightly different in analysis algorithms, our study suggests that RAD1a is the most suitable reference gene for accurate normalization of target gene expression at the developmental stage of the honeybee.
Keywords:
Honeybee, Developmental stage, Reference gene, qRT-PCR, NormalizationINTRODUCTION
The Western honeybee, Apis mellifera L., plays a key pollinator role in natural and agricultural ecosystems (Leonhardt et al., 2013), with an estimated commercial value of over $15 billion per year in the United States (Walsh, 2013). In the Republic of Korea, the economic value of bee pollinating fruit and vegetable agriculture was estimated to be approximately $5 billion (Jung, 2008). In addition to its economic value, the honeybee has been extensively studied as a model insect species due to its highly developed sociability, specialization in division of labor, and ability to manage colonies (Lee and Kim, 2017). Honeybee colonies consist of eggs, larvae, pupae, and adults, and the adults are divided into drones, female workers, and queens (Winston, 1991). The queen bee only consumes royal jelly during the larval stage, and its average development period is usually 16 days which is shorter than that of worker bees (18 days). The queen bee is responsible for oviposition; it can lay as many as 1,500 eggs per day for an average of three to four years of life (Winston, 1991). Unlike the queen bee, which receives royal jelly for the entire larval period, worker bees are developed when larvae are fed regular honey after feeding royal jelly for three days (Asencot and Lensky, 1988). Following emergence, age-related division of labor is based on a form of behavioral development by workers known as age polyethism (Robinson and Huang, 1998). During the first 2~3 weeks of adult life, the nurse bees are typically responsible for in-hive tasks such as brood care (nursing) and hive maintenance. Followed by a transition to tasks outside the hive, forager bees predominantly collect honey and pollen 2~3 weeks before death (Dukas, 2008).
Interestingly, the polyethism of honeybees is not rigid; instead, the honeybee can respond to changes in environment and colony conditions and flexibly alter their typical patterns of age polyethism (Robinson and Huang, 1998). For example, when few nurse bees have remained in the colony, foragers expand their duties to the tasks usually performed by nurses, while in the condition that the number of the forager is limited, nurse bees perform the task of foragers (Robinson and Huang, 1998). Considering the bee colony composing different developmental stages of the honeybee, development of queen/worker by royal jelly feeding, division of nurse/forager, and flexibility of age polyethism, determination of genes putatively involved in bee physiology and investigation of gene expression are essential to extend our knowledge to development and evolution of sociality in insects.
To date, quantitative real-time PCR (qRT-PCR) has been widely suggested as the primary technique for quantifying gene expression levels because of its relatively high speed, sensitivity, reproducibility, and accuracy (Ling and Salvaterra, 2011; Zhai et al., 2014). However, normalization should be performed with reliable reference genes stably expressing across different samples to determine target gene expression levels accurately. Therefore, suitable reference genes should be selected (Wong and Medrano, 2005; Ling and Salvaterra, 2011).
Therefore, in order to study honeybee physiology at different developmental stages, we investigated the expression stabilities of housekeeping genes in different developmental stages (egg, 1st instar larva, 3rd instar larva, 5th instar larva, pupa, nurse, and forager) to suggest appropriate reference genes for gene expression study using qRT-PCR. In the present study, we selected five candidate reference genes, including ribosomal protein S5 (RPS5), ribosomal protein S18 (RPS18), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), ADP-ribosylation factor 1 (ARF1), and Ras-related protein Rab-1A (RAD1a), which have been previously suggested as reference genes in honeybee studies (Moon et al., 2018; Jeon et al., 2020; Kim et al., 2021). However, these genes have not been evaluated in the differenet developmental stages in honeybee, therefore, the stabilities of these five genes across developmental stages were evaluated with Cq distribution analysis and three programs (geNorm, NormFinder, and BestKeeper).
MATERIALS AND METHODS
1. Insects sample preparation and total RNA extraction
The honeybee, A. mellifera (an Italian hybrid), colony used in this study was maintained at an experimental apiary (36°36′69.09″ N, 128°11′70.42″ E) in Sangju-si, Gyeong-sangbuk-do, Republic of Korea. For expression profiles of candidate reference genes across different developmental stages, we collected egges, three larval stages (1st instar, 3rd instar, and 5th instar), pupa, and two adult stages (nurse and forager). For RNA extraction, five eggs, ten 1st instar larvae, five for 3rd instar larvae, and three 5th instar larvae were prepared, and one specimen was used for pupae, nurse, and forager stages. Thus, each sample was collected from five honeybee colonies for five biological replication. Collected samples were immediately frozen with dry ice and stored at -80℃ until RNA extraction.
Each sample was completely homogenized using a bullet blender (Bertin Technologies, Montigny-le-Bretonneux, France), and total RNA was extracted with the yesRTM total RNA extraction kit with a gDNA Eliminator column (Prefilter PF02) (GenesGen, Busan, Korea). The purity and quantity of the extracted RNA were measured in triplicate using a SpectraMax QuickDrop spectrophotometer (Molecular Devices, CA, USA). The prepared RNA was then stored at -80℃ until further use.
2. Primer design and cloning
Candidate reference genes and their primers’ information were obtained from previous studies (Moon et al., 2018; Jeon et al., 2020; Kim et al., 2021). Among five candidate genes, the primer set for RPS18 was designed on two different exons to amplify genomic DNA containing an intron region, thereby amplifying two products if the sample is contaminated with genomic DNA. Amplification specificity and genomic DNA contamination (for RPS18) were determined by visualization on 2% agarose gel and melting curve analyzed by real-time PCR reaction.
For subcloning of candidate reference genes, honeybee total RNA was used as a reverse transcription PCR reaction template using the DiaStarTM OneStep RT-PCR kit (SolGent, Daejeon, Korea). Reaction conditions were as follows: 30 min at 50℃ followed by 15 min at 95℃ (20 sec at 95℃, 40 sec at 56℃, 30 sec at 72℃)×35 cycles and 5 min at 72℃, with each gene-specific primers for amplification (Table 1). The amplicons were purified with the QIAqick PCR purification kit (Qiagen, Valencia, CA, USA) and then subcloned into pGEM®-T easy vector (Promega, Madison, MU, USA). Next, the subcloned plasmid was transformed into DH5α chemically competent Escherichia coli (Ezynomics, Daejeon, Korea). The purified plasmid containing positive inserts was submitted for sequencing reactions using the M13 universal primer with an ABI PRISM 3730XL analyzer (Macrogen, Seoul, Korea).
3. Quantitative real-time PCR
One microgram of RNA extracted from different developmental stages was primed with oligo (dT), and cDNA was synthesized with ReverTra Ace reverse transcriptase according to the manufacturer’s protocol (Toyobo, Osaka, Japan). For qRT-PCR analysis, we used the CFX Connect Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA) with the SYBR GREEN methodology. The PCR efficiency of each gene primer was calculated from the given slope after running a standard curve generated with 3 points of 10-fold serial dilutions of cDNA using the E=10-1/slope formula. Each sample was analyzed in duplicate (technical replicates) in 20 μL of a total reaction volume containing 10 pmol of each primer, 2× Thunderbird SYBR qPCR Master Mix (Toyobo), and 100 ng of cDNA. The PCRs were performed at 95℃ for 1 min, followed by (95℃ for 15 sec, 56℃ for 15 sec, and 72℃ for 30 sec)×40 cycles. Quantification cycle (Cq) values were determined at same fluorescence threshold line (0.1) for each gene.
4. Data analysis
Three software programs were used in this study: geNorm (version 3.2) (Vandesompele et al., 2002), NormFinder (version 0.953) (Andersen et al., 2004), and BestKeeper (version 1) (Pfaffl et al., 2004) to evaluate the expression stability of five candidate reference genes across developmental stages of the honeybee. In addition, we analyzed the Cq distribution of genes with SigmaPlot 14.0, and the arithmetic means (AM), standard deviation (SD), and coefficient of variation (CV) values were calculated (CV=SD/AM). To analyze the Cq distribution of genes, Cq values according to the developmental stages of honeybee were statistically analyzed with one-way ANOVA using SPSS for Windows version 23.0 (IBM, Armond, NY, USA). The geNorm calculates the expression stability value (M) of each putative reference gene based on the geometric mean, and the gene with the lowest M value is the most stably expressed gene in the geNorm analysis. In addition, geNorm determines the average pairwise variation (V) of a gene to estimate the optimal number of reference genes for accurate normalization of target genes (Hellemans et al., 2007). NormFinder calculates the stability value of each candidate gene by overall variation of the gene, wherein the genes showing lower stability values indicate more stable (Andersen et al., 2004). BestKeeper calculates the geometric mean of the Cq values of the genes and then calculates the more stable genes with lower SD values, yielding a suitable reference gene (Pfaffl et al., 2004).
RESULTS
1. Amplification specificity and efficiency
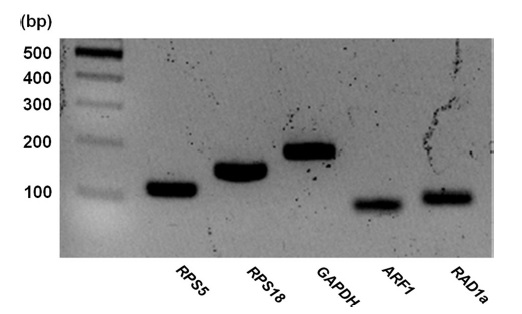
Before qRT-PCR analysis, amplification specificity and efficacy were investigated. All PCR products amplified with the primer set of each reference gene showed a single band on a 2% agarose gel (Fig. 1), and a single peak was detected with melting temperature analysis by RT-PCR (data not shown). In addition, a single band on agarose gel and a single peak in melting curve analysis of RPS18 confirmed no genomic DNA contamination. According to the analysis of PCR efficiencies, all five candidate genes had linear regression coefficients R2>0.997, and amplification efficiencies ranged from 92 to 107% (Table 1).
2. Cq distributions patterns of reference genes
The expression patterns of five candidate reference genes were investigated in the sequence of seven developmental stages of the honeybee. The Cq values among genes in different samples showed variable ranges between 16.18±0.221 (GAPDH in nurse) representing the minimum value and 21.98±0.256 (ARF1 in pupa) representing the maximum value. In the analysis of Cq values of each gene in developmental stages of the honeybee, Cq of RPS5 and RPS18 exhibited similar trends; lowest in 1st instar and highest in forager. In GAPDH, Cq in egg and pupa showed higher than those in other stages, whereas nurses demonstrated the lowest Cq value of GAPDH. The transcription level of ARF1 in 3rd instar larva was the highest, while that in the pupa was the lowest. When the highest and lowest Cq values of RPS5, RPS18, GAPDH, and ARF1 were statistically compared, the expression levels of these genes were significantly different (p<0.05). In addition, Cq values of these four genes in seven developmental stages mainly exhibited statistically varied trends (p=0.000). In contrast, the highest and lowest Cq values of RAD1a were not significantly different, and RAD1a also showed generally similar Cq values across all seven developmental stages of honeybee (Fig. 2A). These results suggest that RAD1a, rather than RPS5, RPS18, GAPDH, and ARF1, can be used as possible reference genes due to its low variation of Cq value.
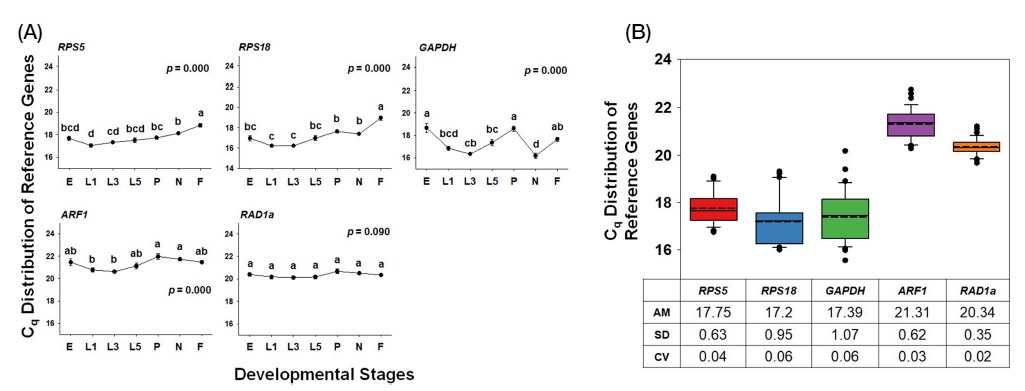
Expression patterns of five candidate reference genes in seven developmental stages of the honeybee. The quantification cycle (Cq) values of candidates were obtained from developmental stages, including egg (E), 1st instar larva (L1), 3rd instar larva (L3), 5th instar larva (L5), pupa (P), nurse bee (N), and forager bee (F). The Cq values were statistically analyzed with One-way ANOVA (A). The Cq values of each gene across different developmental stages were combined, and their distributions were analyzed with box plot comparisons. The horizontal lines in the box indicate the 25th, 50th, and 75th percentile values. The dotted lines in the big box show the mean median. The error bars denote the maximum and minimum values (B).
When Cq values of each gene across different samples were combined, and the values of AM, SD, and CV of the gene were calculated, SD values of GAPDH and RPS18 were 1.07 and 0.95, respectively, and their CVs were 0.06, which were higher than other genes, indicating that these two genes showed high variability of their expression in different developmental stages. In RPS5 and ARF1, SD (0.63 and 0.62, respectively) and CV (0.04 and 0.03, respectively) values were similar. In contrast, RAD1a showed the lowest variability as assessed by low SD (0.35) and CV (0.02) values, indicating that RAD1a is stably expressed across different developmental stages of honeybees (Fig. 2B).
3. Expression stability analysis with three programs
The average expression stability values (M values) were calculated by the geNorm program for each of the five reference genes (Fig. 3). The M value≤0.5 has been proposed as a criterion for selecting an appropriate reference gene (Hellemans et al., 2007; Liu et al., 2014). In the Cq values of candidate genes in seven developmental stages of honeybees, the M values of RPS5 (M=0.46), RAD1a (M=0.431,) and ARF1 (M=0.426) were ≤0.5, whereas those of GAPDH and RPS18 were ≥0.5. Based on the M values analyzed by geNorm, RPS5, RAD1a, and ARF1 are possibly suggested to be suitable reference genes for qRT-PCR in different developmental stages of the honeybee (Fig. 3A).
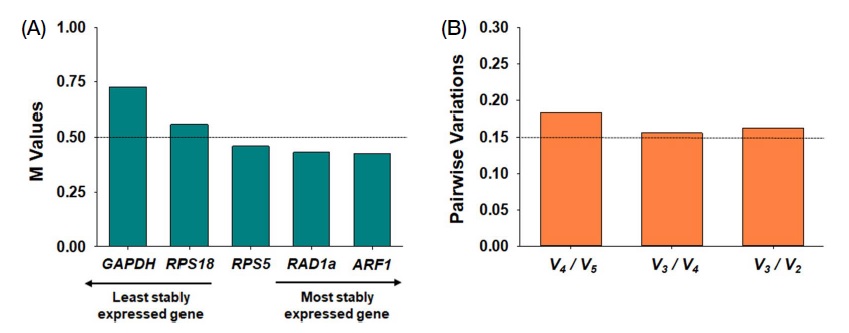
The expression stabilities (M values) and pairwise variation analysis of five candidate reference genes analyzed by geNorm. The dotted line indicates the cutoff M value for the appropriate reference gene selection (A). Pairwise variation analysis was conducted to determine the optimal number of reference genes. The dotted line indicates the cutoff value for the suggestion of an optimal number of reference genes (B).
In addition, pairwise variation (Vn/Vn+1) values representing the optimal number of reference genes for accurate normalization were calculated via geNorm. Previous studies have shown that a value of 0.15 is an appropriate cutoff value in pairwise variation analysis (Vandesompele et al., 2002). However, our candidate reference gene combination exceeded the 0.15 value in all instances (V2/V3, 0.163; V3/V4, 0.156; V4/V5, 0.183), indicating that a combination of multiple reference genes is not optimal for target gene normalization. However, using a single reference gene might be appropriate for qRT-PCR study in honeybee developmental stages (Fig. 3B).
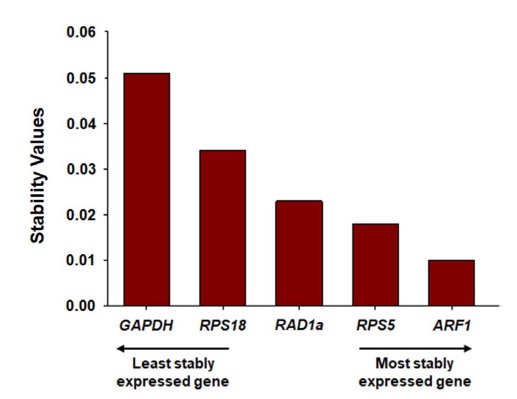
We operated the NormFinder program to identify optimal reference genes by calculating stability values based on the expression variation of candidate genes (Andersen et al., 2004; Liu et al., 2014). According to the average stability values arithmetically calculated according to the developmental stages of the honeybee, ARF1 was found to be the most stable gene (average value=0.010). The stability ranking from the most stable (lowest stability value) to the least stable (highest stability value) gene was ARF1 (0.010)>RPS5 (0.018)>RAD1a (0.023)>RPS18 (0.034)>GAPDH (0.051) in all samples (Fig. 4).
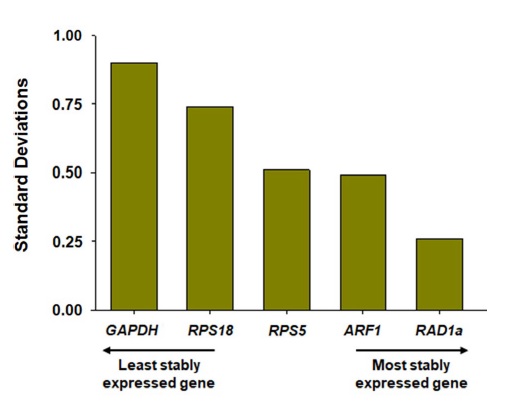
BestKeeper revealed the expression stabilities of candidate reference genes as judged by the inspection of SD values. According to the SD values of five genes in this study, GAPDH and RAD1a were the least stable and the most stable genes, respectively, due to their respective highest and lowest SD values, while ARF1, RPS5, and RPS18 resulted in the second, third, and fourth stable genes. A wide distribution of Cq values of genes as indicated by SD and CV value (Fig. 2B) seemed to yield a higher SD value analyzed by BestKeeper (Fig. 5).
DISCUSSION
Considering that the honeybee is a good model insect for molecular physiological studies of social development (Consortium, 2006), determining the expression level of genes putatively involved in honeybee physiology is essential. In addition, appropriate reference genes stably expressing across the different sample conditions should be selected for accurate normalization of the gene of interest with qRT-PCR (Lourenço et al., 2008; Scharlaken et al., 2008; Reim et al., 2013). Therefore, several studies have been conducted to evaluate qRT-PCR reference genes in honeybees (Lourenço et al., 2008; Scharlaken et al., 2008; Reim et al., 2013; Moon et al., 2018; Jeon et al., 2020; Kim et al., 2021).
In this study, in order to select suitable reference genes in different developmental stages of the honeybee, we evaluated the expression stability of five candidate reference genes (RPS5, RPS18, GAPDH, ARF1, and RAD1a) across seven developmental stages (egg, 1st instar larvae, 3rd instar larvae, 5th instar larvae, pupa, nurse, and forager) by analyzing Cq distribution of the genes and three analysis algorithms programs (geNorm, NormFinder, and BestKeeper). Although the selected five candidates have not been evaluated in different developmental stages, they were previously suggested as the appropriate reference genes in the honeybee. According to the previous studies, RPS5, RPS18, and GAPDH represented optimal reference genes in various tissue of honeybee collected across seasons (Jeon et al., 2020). In addition, RPS18 and GAPDH were suggested to be employed as suitable reference genes for qRT-PCR-based determination of seasonal and labor-specific gene expression profiles (Moon et al., 2018). Furthermore, RAD1a and ARF1 were suggested to be the reliable reference genes in different tissues and two adult stages (nurse and forager) of honeybees exposed to various pesticides (Kim et al., 2021). The selection of these five candidates was reliable because they mostly show stable expression regarding their cellular physiological function. The ribosomal proteins (RPS5 and RPS18) and metabolic enzyme (GAPDH) are essentail housekeeping genes involved in protein expression (Zhou et al., 2015) and glycolysis (Tunio et al., 2010), respectively. Two genes (ARF1 and RAD1a) belong to the Ras superfamily and are responsible for intracellular and membrane transport (Chavrier and Goud, 1999).
According to the results analyzed by geNorm, NormFinder, and BestKeeper, the three algorithms revealed slightly different stability values of the five candidate reference genes, as observed in previous studies (Moon et al., 2018; Wang et al., 2019; Jeon et al., 2020; Kim et al., 2020); therefore, the combined use of results analyzed by all programs would ensure more credible suggestion for optimal reference genes (Wang et al., 2019; Jeon et al., 2020). In the analysis of NormFinder, the NormFinder program does not provide an appropriate cutoff value, but recent studies suggested that a suitable cutoff value of gene expression stability is ≤0.15 (McMillan and Pereg, 2014; Julian et al., 2016). Based on these criteria, all five genes met the values, implying that RPS5, RPS18, GAPDH, ARF1, and RAD1a would be possibly suggested to be applied as the qRT-PCR reference genes (Fig. 4). However, considering M≤0.5 as a criterion for selection of an appropriate reference gene in the geNorm analysis (Hellemans et al., 2007; Liu et al., 2014), geNorm resulted in three genes (RPS5, RAD1a, and ARF1) as optimal reference genes (Fig. 3A). Although the rankings of the stability values of these three genes were different between geNorm and NormFinder analysis, RPS5, RAD1a, and ARF1 were also ranked in the high stable genes when compared with GAPDH and RPS18 in NormFinder (Fig. 4). Furthermore, BestKeeper revealed that RAD1a, ARF1, and RPS5 were the first to third stable genes, respectively, as judged by their lower SD values than RPS18 and GAPDH, although SD values of all five genes were ≤1.0, which is the cutoff line in BestKeeper analysis (Fig. 5) (Chechi et al., 2012). Likewise, Cq distribution analysis demonstrated that RAD1a, ARF1, and RPS5 exhibited relatively lower CV values than RPS18 and GAPDH (Fig. 2B), suggesting that RAD1a, ARF1 and RPS5 are suggested to be applicable as optimal reference genes in different developmental stages of the honeybee. However, when each Cq value of RAD1a, ARF1, and RPS5 in seven different developmental stages was compared, the highest Cq and lowest Cq of RPS5 and ARF1 were significantly different (p<0.05). In contrast, RAD1a showed statistically similar expression levels across developmental stages from egg to forager (p>0.05) (Fig. 2A), supporting that RAD1a is the most optimal reference gene for normalization of target gene using qRT-PCR in honeybee developmental stage.
In the present study, pairwise variations were also analyzed by geNorm to suggest an optimal number of reference genes for target gene normalization. According to the previous studies, Vn/Vn+1≤0.15 is the criterion (Vandesompele et al., 2002; Wan et al., 2010), but the value of pairwise variation for a combination of multiple reference genes were higher than 0.15 in this study (Fig. 3B). This indicates that the addition of the reference gene does not significantly contribute to the accurate normalization of the target gene in honeybee developmental stages, as suggested in the previous study (Silveira et al., 2009). A single gene, particularly RAD1a, showed the most negligible variation of Cq across different developmental stages (Fig. 2) and SD value in BestKeeper (Fig. 5). In addition, the M and stability value of RAD1a analyzed by geNorm (Fig. 3A) and NormFinder (Fig. 4) were under cutoff lines, suggesting that a single use of RAD1a as a reference gene can be appropriated in the gene expression study of the developmental stages of honeybee.
In conclusion, we evaluated the expression stabilities of five candidate reference genes in different developmental stages of the honeybee with Cq distribution analysis and three programs. Considering the cutoff line of CV≤0.5 for Cq distribution (Fig. 2B), stability value≤0.15 for NormFinder (Fig. 4), and SD≤1.0 for BestKeeper (Fig. 5), all five genes were applicable as the reference genes. However, in geNorm analysis, three genes (RPS5, RAD1a, and ARF1) were suggested as reference genes because their M values were ≤0.5 (Fig. 3A), but Cq values of RAD1a, rather than RPS5 and ARF1, were statistically similar across seven developmental stages (Fig. 2A), suggesting that RAD1a is suitable reference gene. Pairwise variation analysis further suggested a single use of the reference gene (Fig. 3B). Therefore, in this study, we finally suggest that RAD1a is the most appropriate reference gene for the accurate normalization of target gene expression at the developmental stage of the honeybee, but selected reference gene is remained to be validated by normalization of target gene.
Acknowledgments
This study was supported by a research fund (PJ015778012021) from Rural Development Administration.
References
-
Andersen, C. L., J. L. Jensen and T. F. Ørntoft. 2004. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64: 5245-5250.
[https://doi.org/10.1158/0008-5472.CAN-04-0496]

-
Asencot, M. and Y. Lensky. 1988. The effect of soluble sugars in stored royal jelly on the differentiation of female honeybee (Apis mellifera L.) larvae to queens. Insect Biochem. 18: 127-133.
[https://doi.org/10.1016/0020-1790(88)90016-9]

-
Chavrier, P. and B. Goud. 1999. The role of ARF and Rab GTPases in membrane transport. Curr. Opin. Cell Biol. 11: 466-475.
[https://doi.org/10.1016/S0955-0674(99)80067-2]

-
Chechi, K., Y. Gelinas, P. Mathieu, Y. Deshaies and D. Richard. 2012. Validation of reference genes for the relative quantification of gene expression in human epicardial adipose tissue. PLoS One 7: e32265.
[https://doi.org/10.1371/journal.pone.0032265]

-
Consortium, H. G. S. 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443: 931-949.
[https://doi.org/10.1038/nature05260]

-
Dukas, R. 2008. Mortality rates of honey bees in the wild. Insectes Soc. 55: 252-255.
[https://doi.org/10.1007/s00040-008-0995-4]

-
Hellemans, J., G. Mortier, A. De Paepe, F. Speleman and J. Vandesompele. 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8: 1-14.
[https://doi.org/10.1186/gb-2007-8-2-r19]

-
Jeon, J. H., K. Moon, Y. Kim and Y. H. Kim. 2020. Reference gene selection for qRT-PCR analysis of season-and tissue-specific gene expression profiles in the honey bee Apis mellifera. Sci. Rep. 10: 1-13.
[https://doi.org/10.1038/s41598-020-70965-4]

-
Julian, G. S., R. W. de Oliveira, S. Tufik and J. R. Chagas. 2016. Analysis of the stability of housekeeping gene expression in the left cardiac ventricle of rats submitted to chronic intermittent hypoxia. J. Bras. Pneumol. 42: 211-214.
[https://doi.org/10.1590/S1806-37562015000000133]

- Jung, C. 2008. Economic value of honeybee pollination on major fruit and vegetable crops in Korea. J. Apic. 23(2): 147-152.
-
Kim, S., S. Cho and S. H. Lee. 2021. Selection of stable reference genes for real-time quantitative PCR in honey bee pesticide toxicity studies. J. Apic. Res.: 1-11 (in press).
[https://doi.org/10.1080/00218839.2021.1950972]

-
Kim, Y., Y. Kim and Y. H. Kim. 2020. Evaluation of reference genes for gene expression studies using quantitative real-time PCR in Drosophila melanogaster after chemical exposures. J. Asia Pac. Entomol. 23: 385-394.
[https://doi.org/10.1016/j.aspen.2020.01.008]

-
Lee, S. H. and Y. H. Kim. 2017. Comparative proteome analysis of honey bee workers between overwintering and brood-rearing seasons. J. Asia Pac. Entomol. 20: 984-995.
[https://doi.org/10.1016/j.aspen.2017.07.011]

-
Leonhardt, S. D., N. Gallai, L. A. Garibaldi, M. Kuhlmann and A. M. Klein. 2013. Economic gain, stability of pollination and bee diversity decrease from southern to northern Europe. Basic Appl. Ecol. 14: 461-471.
[https://doi.org/10.1016/j.baae.2013.06.003]

-
Ling, D. and P. M. Salvaterra. 2011. Robust RT-qPCR data normalization: validation and selection of internal reference genes during post-experimental data analysis. PLoS One 6: e17762.
[https://doi.org/10.1371/journal.pone.0017762]

-
Liu, C., N. Xin, Y. Zhai, L. Jiang, J. Zhai, Q. Zhang and J. Qi. 2014. Reference gene selection for quantitative real-time RT-PCR normalization in the half-smooth tongue sole (Cynoglossus semilaevis) at different developmental stages, in various tissue types and on exposure to chemicals. PLoS One 9: e91715.
[https://doi.org/10.1371/journal.pone.0091715]

- Lourenço, A. P., A. Mackert, A. dos Santos Cristino and Z. L. P. Simões. 2008. Validation of reference genes for gene expression studies in the honey bee, Apis mellifera, by quantitative real-time RT-PCR. Apidologie 39: 372-385.
-
McMillan, M. and L. Pereg. 2014. Evaluation of reference genes for gene expression analysis using quantitative RT-PCR in Azospirillum brasilense. PLoS One 9: e98162.
[https://doi.org/10.1371/journal.pone.0098162]

-
Moon, K., S. H. Lee and Y. H. Kim. 2018. Validation of quantitative real-time PCR reference genes for the determination of seasonal and labor-specific gene expression profiles in the head of Western honey bee, Apis mellifera. PLoS One 13: e0200369.
[https://doi.org/10.1371/journal.pone.0200369]

- Pfaffl, M. W., A. Tichopad, C. Prgomet and T. P. Neuvians. 2004. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26: 509-515.
-
Reim, T., M. Thamm, D. Rolke, W. Blenau and R. Scheiner. 2013. Suitability of three common reference genes for quantitative real-time PCR in honey bees. Apidologie 44: 342-350.
[https://doi.org/10.1007/s13592-012-0184-3]

-
Robinson, G. E. and Z.-Y. Huang. 1998. Colony integration in honey bees: genetic, endocrine and social control of division of labor. Apidologie 29: 159-170.
[https://doi.org/10.1051/apido:19980109]

-
Scharlaken, B., D. C. de Graaf, K. Goossens, M. Brunain, L. J. Peelman and F. J. Jacobs. 2008. Reference gene selection for insect expression studies using quantitative realtime PCR: The head of the honeybee, Apis mellifera, after a bacterial challenge. J. Insect Sci. 8: 1-10.
[https://doi.org/10.1673/031.008.3301]

-
Silveira, É. D., M. Alves-Ferreira, L. A. Guimarães, F. R. da Silva and V. T. de Campos Carneiro. 2009. Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biol. 9: 1-10.
[https://doi.org/10.1186/1471-2229-9-84]

-
Tunio, S. A., N. J. Oldfield, D. A. Ala’Aldeen, K. G. Wooldridge and D. P. Turner. 2010. The role of glyceraldehyde 3-phosphate dehydrogenase (GapA-1) in Neisseria meningitidis adherence to human cells. BMC Microbiol. 10: 1-10.
[https://doi.org/10.1186/1471-2180-10-280]

-
Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3: 1-12.
[https://doi.org/10.1186/gb-2002-3-7-research0034]

- Walsh, B. 2013. The plight of the honeybee. Time Mag. 182: 31-37.
-
Wan, H., Z. Zhao, C. Qian, Y. Sui, A. A. Malik and J. Chen. 2010. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal. Biochem. 399: 257-261.
[https://doi.org/10.1016/j.ab.2009.12.008]

-
Wang, Z., Q. Meng, X. Zhu, S. Sun, S. Gao, Y. Gou and A. Liu. 2019. Evaluation and validation of reference genes for quantitative real-time PCR in Helopeltis theivora Waterhouse (Hemiptera: Miridae). Sci. Rep. 9: 1-10.
[https://doi.org/10.1038/s41598-019-49479-1]

- Winston, M. L. 1991 The biology of the honey bee. 300p. Harvard university press, Cambridge, Massachusetts.
-
Wong, M. L. and J. F. Medrano. 2005. Real-time PCR for mRNA quantitation. Biotechniques 39: 75-85.
[https://doi.org/10.2144/05391RV01]

-
Zhai, Y., Q. Lin, X. Zhou, X. Zhang, T. Liu and Y. Yu. 2014. Identification and validation of reference genes for quantitative real-time PCR in Drosophila suzukii (Diptera: Drosophilidae). PLoS One 9: e106800.
[https://doi.org/10.1371/journal.pone.0106800]

-
Zhou, X., W.-J. Liao, J.-M. Liao, P. Liao and H. Lu. 2015. Ribosomal proteins: functions beyond the ribosome. J. Mol. Cell Biol. 7: 92-104.
[https://doi.org/10.1093/jmcb/mjv014]