Validation of Antioxidant and Immune Related Gene Members among Apis cerana Species of South India by Quantitative Real-Time PCR
Abstract
Apis cerana most widely found bee species of southern parts of India is now at stress. Due to the increasing bee cultivation centres, these bees are under tremendous stress levels from both external and internal agents. External agents might be due to constant application of miticides and other chemical compounds. Wild varieties which are in a free setup, though infected by Varroa mites, could face little pressure. We designed to study the effect of the habitat settings (natural and captive) on the bee species. Natural ones are those which are seen in forests, trees and other house roofs. Captive are those seen among the bee keeping centres. Antioxidant and immune related gene members were studied for their expression levels between the natural and captive environments. As hypothesized, we found significant effects on the expression of the gene members. Bees kept in captivity showed over expression of both antioxidant and immune related genes when compared to the natural bees. Ribosomal protein (RP4) was used as housekeeping gene throughout the study. Among them 2 members, CYP6AS3 and CYP6AS10 showed contradictory results. Their expression was little more in natural bees when compared to the captive bees. This study could pave ways of how to effectively manage the bees among the cultivated centres so as to get more yields, keeping in view of the colony collapse disorder. This validation studies could also throw knowledge on the actual mechanism involved in the stress regulation among this sweet little bees.
Keywords:
Apis cerana, Antioxidant genes, CYP6AS3 and CYP6AS10, Natural vs CaptiveINTRODUCTION
In artificial and natural habitat, pollination of plants by honeybees (Apis mellifera) not only contributes for production of food and nourishment but also leads to increase in biodiversity (Li, 2015). From the reports, it is estimated that globally, there is an overall increase in colony loss and honey bee depopulation. Apart from pathogens (parasites, fungi, bacteria and viruses), environmental factors, agrochemical use, many other factors seems to be reasons for the decline in honeybee population, which eventually is altering the immune system of bee for defense mechanisms.
Honey bees defend themselves against infectious and parasitic organisms by the innate immune system which also includes physical barriers and comprehensive cellular and humoral responses. Overall health status and immune system of bees can be affected by pathogens, acaricides, fungicides, herbicides and other pesticides. Pathogen recognition receptors, innate immune system effectors and signalling pathways are those defense mechanisms associated with bee immune system.
To control Varroa mite (Johnson et al., 2010) treatment by miticides have been introduced as management strategy. Honey bee products were contaminated due to irrelevant application of miticides and this not only inflicted undesirable effects on the health of bees but also imparted the pesticide tolerance of Varroa mites (Johnson et al., 2013). Harmonious color, aroma, and structure of honey bees and biological characteristics are of great importance because of endured relationship and coevolution with plants as pollinators in natural habitats (Garibaldi et al., 2013). Considerably there was increase in fruit seed rate and yield as the bees were introduced into greenhouses for pollination which led to reduction in the cost of artificially assisted pollination. However, there is a negative impact on the foraging behaviour and reproduction of bees due to reduced area resulting in variation of temperature and humidity (Ollerton et al., 2012).
1. Antioxidant system in Apis cerana
By-products of aerobic metabolism generate Reactive Oxygen Species (ROS) frequently. The data collected on ROS suggest that it causes oxidative damage to cellular components and responsible for retrogressive diseases and ageing. Mitochondria generate ROS which consume about 90% of the oxygen produced by the cell (Perez-Campo et al., 1998) where primary and secondary antioxidant enzymes present in the cell act directly or indirectly on ROS molecules (Valko, 2006). ROS is attacked by first line of defence, with three types of primary antioxidant enzymes like (1) superoxide dismutases (SODs) which rearranges the superoxide to oxygen and hydrogen peroxide (2) Catalase, which aids in breaking down of hydrogen peroxide into oxygen and water further preventing free hydroxyl radical formation and lastly (3) Peroxidases, which catalyses analogous reaction by reducing hydrogen peroxide to water acting as an electron donor, normally reduced thioredoxin (TRX) or glutathione (GSH) (Abele et al., 2002). The activity of Catalase (CAT), glutathione S-transferase (GST) and superoxide dismutase (SOD) was determined in postmitochondrial fractions of tissue homogenates (spermathecae, muscle and ventriculi), in hemolymph plasma, and in semen of honey bees (Comhair, 2005).
Gene which is responsible for bacterial virulence and stress tolerance in variable atmosphere was found to be Methionine sulphoxide reductase A msrA (Douglas, 2004). Downstream activation of ERK (MAPK1/ERK2 and MAPK3/ERK1) signalling by an enzyme Serine/threonine-protein kinase intervenes mitogenic and stress-induced activation of transcription factors, regulation of translation, and mediation of cellular proliferation, survival rate and differentiation (Li et al., 2019). For example oxidative stress in most of the microorganisms is protected by the significant gene MsrA, but it is not proved that oxidative stress inducible gene is been present in such bacteria. The mechanism of regulation of msrA expression and the physiological function(s) of the gene product in most of the microorganisms is not yet clearly explained (Tamburro, 2001).
2. Immune related stress genes
It was suggested that stress response with increased expression level under brood rearing - suppressed condition is because of enzyme acetyl cholinesterase 1 (AmAChE1) of honey bee (Kim et al., 2019). According to the recent studies, the state of brood rearing increased when suppression of brood rearing occurred and their correlation was estimated by the expression of honey bee acetylcholinesterase1 (AmAChE1) (Kim, 2017). Honey bee colonies suffered stressful condition due to suppression of brood rearing assumed that stress response or stress management involved AmAChE1 as suggested by some of the scientists. In addition to bees, pinewood nematode (Bursaphelenchus xylophilus) and fruit fly (Drosophila melanogaster) produced soluble AChE they also suffered from chemical stress. So it was finalised that AmAChE1is factor for stress regulation pathway in bees (Kim, 2014). Resistance in the insects from xenobiotic toxins administered by most important mechanisms were called metabolic detoxification. Such metabolic detoxification was neutralized by detoxification enzyme systems like Cytochrome P450 monooxygenases (CYPs, also called P450s), carboxylesterases (COEs) and glutathione S-transferases (GSTs) (Berenbaum and Johnson, 2015).
The primary metabolite, P450 enzymes in honey bee which detoxify the pesticides and secondary metabolites from plants were reported in the performed experiments. Pyrethroid insecticides and neonicotinoid insecticides on honey bees could not enhance the toxicity due to the presence of P450 inhibitor piperonyl butoxide (PBO) (Johnson et al., 2006). Mao et al. (2017) discovered that detoxification of dietary phytochemicals by P450 was inhibited by myclobutanil, led to the intrusion of ATP production and inadequate amount of energy for flight and other actions. The metabolism of quercetin, a flavonoid component of honey and pollen include the involvement of CYP6AS1, CYP6AS3, CYP6SA4 and CYP6AS10 of the CYP6 subfamily (Mao et al., 2009). Redox reactions in cell are catalyzed by a wide spectrum of powerful disulfide oxido-reductase, Thioredoxin (TRX). Research on utilization of transcriptomics and proteomics and the role of TRX and TR was initiated (Jobin et al., 1999). During stress conditions like heat and hydrogen peroxide stress, gene trxA was induced in Bacillus subtilis and Oenococcus oeni (Serrano, 2007).
To confirm the differential expression patterns of the genes and assess their utility as potential biomarkers for selecting Varroa tolerant colony phenotypes, the present study analyzed the differential expression patterns of ten genes selected from previous DNA microarray (Jiang et al., 2016) by using real time quantitative PCR in a wide range of honey bee colonies with varying degrees of Varroa tolerance to Varroa parasitism. The effects of three commonly used miticides (Apistan®, Apivar®, Thymovar®) on the expression of three potential biomarker genes were also investigated. In addition, one of the potential biomarker genes, AmCbE E4 encoding a putative esterase E4, was further heterologously expressed and functionally characterized in E. coli (Schöning, 2012).
Hence, identification and screening for the upregulation or downregulation of these gene stress related gene members that underlie the tolerant mechanism would be very much useful in saving this little bees from excessive external pressures. The study was designed to evaluate how chronic exposure to captive conditions could induce physiological stress on bees and thereby regulating antioxidant and immune related genes through quantitative real time expression (qPCR) (Weirich et al., 2002).
MATERIALS AND METHODS
Sample Collection & processing: Apis cerana species samples were collected from different geographical and ecological areas of south India mentioned in the above context from June 2017~September 2019 Samples from both the managed and wild or natural hives were collected from the sample area. In total about 20 samples were taken from each area. Local people who are experienced in handling bee hives and bees were assisting us throughout the study in collecting the samples. Feral colonies were located basing on the movements of the bees towards the nest entrance as mentioned by Sheetal and Basavarajappa (2009). In case of collecting the samples from wild, vegetation and nesting sites seems non uniform owing to different geographic and ecological zones. Field trips were done to such areas which includes scrubby vegetation, rocky areas, agricultural lands, avenue trees, trees in the cultivable lands, buildings of residential area, old mud houses.
1. Sampling sites
- (1) Kerala: Mallappally, Mavelikara, Erumeli, Pandalam, Puliyoor, Kanjiramattom, Chingavanam, Ettumanoor, Evoor, Kalpetta, Nooranadu, Kumbanad, Kaviyoor, Vagathanam, Chenganoor, Kanjirappally and Pathanapuram.
- (2) Andhra Pradesh: Kasipatnam, Rhummanavalsa, Damuku, Anathagir, Thatipudi, Ethamananu valsa, Dhamsarai, Sivalingapuram, Kota padu, Peadhapadu, Borra, Musidipalli, Singada, Augubili, Gadilova, Tennu bodavara, Sunkarameta, ARAKU valley, Dumbriguda, Paderu, Chintapalli, Korukonda.
- (3) Tamil Nadu: Aandiruppu, Kasipatnam, Rhummanavalsa, Aandiruppu, Achampatti, Achanoor, Adambai, Adanakottai, Agaramangudi, Agarathur, Alaginayagipuram, Alamathikkadu, Alathur, Ichankottai, Illupaividuthy, Anathukanpatti, Indalur, Irumbuthalai, Jambugapuram, Kalichankkottai, Kallapuliyur, Kallur, Kalugapulikkadu, Kalyanaodai, Kambaranatham, Chakkarapalli, Chellampatti, Cholapuram.
- (4) Karnataka: Bakkala, Jaddigadde, Vanalli, Kakkalli, Salkani, Onikere, KoLigar, Hulekal, Sonda, Vajagadde, Heggarni, Siddapura, Nilkunda, Bedkani, Muttige, Goranamane, NaNikatta, Yana, Devimane, Idagundi, Manchikeri, Mavinakatta, Magodu.
Satellite images showing the different geographical regions of the sampling sites. Left: Kerala; Right: Andhra Pradesh. Sampling sites are spotted as red in colour.
Satellite images showing the different geographical regions of the sampling sites. Left: Tamil Nadu; Right: Karnataka. Sampling sites are spotted as red in colour.
About 20~25 bee samples from different regions (both managed and wild) were collected from the hives between 08.00 and 17.00 hours during day time. The samples were labelled accordingly and used for the antioxidant analysis.
2. Antioxidant enzyme activities
Thoracic muscle was dissected from the samples and rinsed in ice-cooled PPT buffer. The tissues were then thoroughly homogenized with 5~8 volumes of PPT buffer in glass micro homogenizers and were spinned down for 20 min at 12,000 rpm (4℃). Following elimination of the debris, supernatant containing the postmitochondrial fractions was further saved for the enzyme assays.
CAT activity was assayed by the method of Meharan et al. (2014). Assay mixtures were made of 10 mM H2O2, and different concentrations of enzymes (in 50 mM potassium phosphate buffer, pH 7.0). Enzyme activities were calculated using 0.0394 mM-1·cm-1 as absorption coefficient at 240 nm. 40 μL of the serially diluted H2O2 was sued as H2O2 standard. 20 μL of 550 μM H2O2 standard was used as Catalase negative control. About 20~40 μL of sample was added to each test well labelled. Following incubation, 20 μL of catalase solution was added to all the wells except negative control. Following 1 hr incubation on the shaking incubator, 135 μL of HRP reagent mixture solution was added and the absorbance was measured at 240 nm using ELISA plate reader (Genetix, Germany).
This was assayed according to the technique described by Meharan et al. (2014). In brief, about 0.75 mL of diluted sample was added with 1 mL of 0.01 M Tris-Cl (pH 8.5) and 0.5 mL of pyrogallol. After mixing, the contents were incubated at 37℃ for 20 min. 1.5 mL of water along with 1 mL of 0.01 M Tris-Cl (pH 8.5) and 0.5 mL of pyrogallol serves as negative control. Following loading the samples onto the respective wells, absorbance was recorded at 420 nm are for 0 and 20 min time intervals. The rate of increase on absorbance units (A) per minute for the blank and for the test sample are calculated by the equation (A420 nm/min=O.D at last interval-O.D at first interval/time in min). The % inhibition is given by the formula (A420 nm/min [blank] -A420 nm/min [test]/A420 nm/min [blank]×100.
The total RNA content from the thorax samples were extracted according to the method described by Williams et al. (2013). The frozen tissues (stored at -80℃) were placed into a disposable extraction bag and added with 500 μL lysis/stabilization solution (94.53 g guanidine thiocyanate, 30.45 g ammonium thiocyanate, 33.4 mL of 3 M sodium acetate, pH 5.5 mL) per bee. The contenst were mashed until homogenized. After settling, 620 μL of extraction liquid was placed into into a pre-labelled 1.5 mL micro tube and added with 380 μL acid phenol. After thorough mixing, contenst were vortexed for 30 sec and incubated for 10 min in a 95℃ hot block. Following which the tubes were transferred from hot block to pre-chilled rack in ice and incubated on ice for 20 min. following incubation, 200 μL chloroform was added and shaken vigorously for 1sec and incubated at RT 3 min. The contents were then centrifuged at 14,000 rpm for 15 min at 4℃, and the 500 μL upper phase was transferred to a fresh tube. To the tube equal volume of isopropanol (100%) was added and invert gently to mix the contents. Following incubation at RT 15 min, the contents were spinned down at 10,000 rpm for 10 min at 4℃. The pellet was now washed with 1 mL of ice cold 75% EtOH and centrifuged at 10,000 rpm for 2 min at 4℃. Resuspend the pellet in 200 μL nuclease-free H2O and store at -80℃.
The cDNA synthesis was carried using the RT PCR kit using SuperScript TMII Reverse Transcriptase, 200 U/μL (HiMedia). In brief, 1.5 μg of the RNA obtained in the previous section was used as the starting reaction. The RNA concentration obtained was about 1.65 μg/μL. Hence, about 1.34 μL of the total RNA was used in the reaction. Random primers and 1 μL of RT enzyme were added mixed thoroughly, and the contents were incubated at 25℃ for 10 min. Followed by the incubation at 70℃ for 15 min, the cDNA obtained was then stored and used for real-time PCR analysis.
Primers for real time (Table 1) were designed using primer3 software and were purchased from Sigma-Aldrich. The real-time PCR assay was then performed according to Deepak et al. using the iQTM SYBR Green Supermix (HiMedia). The primers (600 nM) and 1 μL of the RT products were used in the PCR assay, and the reaction was done in a total volume of 12.5 μL. All the reactions were carried out in duplicates and also run in parallel with its respective negative control, to confirm the positive amplification.
Real-time quantification was carried on all the samples of the four regions in the Corbett Research cycler (Bio-Rad). Antioixdant gene members like Catalase (CAT) [Gene ID: 107993802]; Superoxide dismutase 1 (SOD1) [Gene ID: 108000927]; Methionine sulphoxide reductase A (MsrA) [HQ219725.1]; Thioredoxin reductase (Trxr1) [GU188974.1] were used for the study. Immune related gene members like AChE-1 [XM_017050996.1]; AChE-2 [XM_017066417.2]; CYP6AS1 [XM_017066590.2]; CYP6AS3 [XM_028669828.1]; CYP6AS4 [XM_395671.4]; CYP6AS10 [XM_028669827.1].
1.35 μL of the RNA products were initially used, and the program was run for about 40 cycles at 94℃ for 60 s, 63℃ for 45 s, and with an elongation at 73℃ for 60 s for antioxidant gene expression. The program was run for about 40 cycles at 92℃ for 50 s, 64℃ for 55 s, and with an elongation at 74℃ for 60 s for immune related gene expression. The housekeeping gene Ribosomal protein 39S (Rp4) [XM_017064320.2] (forward: 5′ GCAGTTGCTGCAATGAACACT 3′ and reverse: 5′ TCATAGGCATTGCATCTGGCA 3′; 178 bp) was also amplified along with the respective gene of interest for a comparative analysis of the mRNA expression. The comparative analysis of the relative levels of mRNA of the test samples (both natural and captive) was quantified using ΔΔCt method. The Ct values obtained for the gene of interest were normalized to its housekeeping gene.
The reaction was carried out in a total volume of 12.5 μL. All the reactions were carried out in duplicates. Each sample was also run in parallel with its respective negative control, to confirm the positive amplification. Specific amplification programs were selected, for the assay, and in addition, a melting curve program was also done simultaneously, to avoid the mispairing of primers and primer dimer formation.
All the experiments were done in triplicates and mean of the values was taken. A two way ANOVA was used for analysing the data statistically throughout the study (P<0.05). The values were represented as value±SE.
To comparatively analyse the relative levels of mRNA expression, of the test samples, with the control, a standard method of ΔΔCt method of quantification was used. The software machine, after running the samples, gives out the Ct values, for the respective genes. All the reactions are done usually in duplicates. House keeping gene and negative control are also run in the same program, in parallel to the gene of interest. Ct values, are inversely related to the quantity of the nucleic acid present in the test sample i.e., the high the Ct value, the more the quantity of DNA in sample. In general the Ct values, less than 27 indicate that the reaction is strongly positive. Ct values of 30~37 indicate, a weak reaction, and can be considered as no amplification.
RESULTS
1. CAT assay
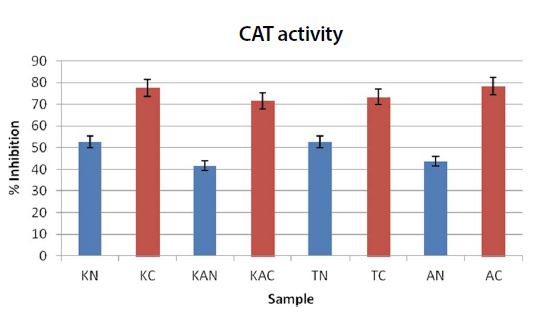
CAT activities seem to be significantly different between the captive and natural samples. There was a significant effect of the CAT activity between the regions and habitats remembered at the P<0.05 level. The significance effect of the CAT activity on the natural and captive [F(19,133)=2.934, P=0.00016] and [F(7,133)=160.25, P=1.33E-61]. However, there was no significant effect seen between the regions, and more or less similar in terms of CAT activity. But the CAT inhibition was found to be significantly higher than the natural settings in all the four regions (Fig. 3). This could be the possible explanation for the stress levels induced within the captive species, on exposure to pressures.
2. Superoxide dismutase (SOD) assay
SOD activities were in accordance to the CAT assay results and also seem to be significantly different between the captive and natural samples. There was a significant effect of the CAT activity between the regions and habitats remembered at the P<0.05 level. The significance effect of the CAT activity on the natural and captive [F(19,133)=1.9384, P=0.015935] and [F(7,133)=123.783, P=4.59E-55]. In contrast, the natural setting samples showed no significance between the regions, but in captivity, the difference was seen between Kerala, Karnataka and Andhra, Tamil Nadu zones. This could possibly be due to the climatic variations or the type of miticides used (Fig. 4). This could be the possible explanation for the stress levels induced within the captive species, on exposure to pressures. From the results showed in both CAT and SOD assay, the enzyme activity in Apis cerana bees of captive habitats of all the four regions of study was higher than the natural settings.
3. Real time PCR
We validated the PCR array data on a larger sample size (80 which includes both captive and natural) using gene-specific qRT-PCR assay. We selected the 10 genes that showed differential expression among the samples in different regions and habitats (CAT, SOD1, MsrA, Trxr1, AChE-1, AChE-2, CYP6AS1, CYP6AS3, CYP6AS4 and CYP6AS10) for the validation experiment.
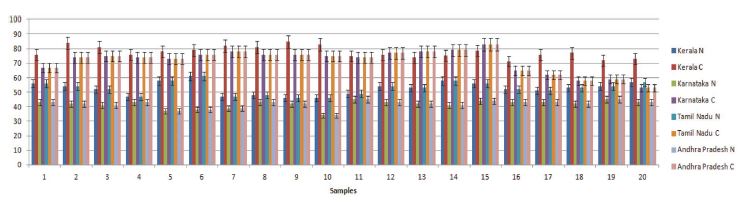
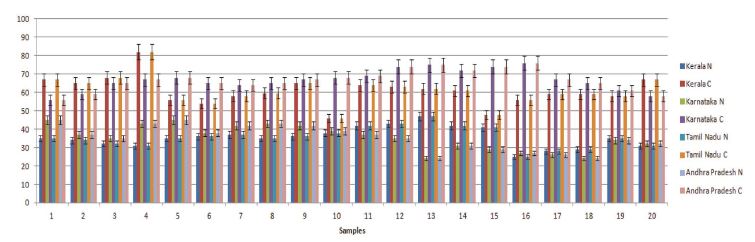
From the quantitative real-time PCR (qPCR) results, it was observed that the stress marker genes used in the study showed varying expression patterns among the samples from different regions (Fig. 1). When samples of natural settings from the 4 geographical regions were compared to the captive settings, captive groups showed higher expression levels of the above genes. All of the genes expression values were significantly higher than those of natural setting except CYP6AS3 (P=0.009) and CYP6AS10 (P=0.049).
From the real time PCR data it was obvious that captive species showed upregulation among the antioxidant and immune related gene members between all the study regions. But among the ten two members CYP6AS3 and CYP6AS10 showed no significance among the natural and captive. In Andhra and Karnataka regions there is significant effect found between the natural and captive. In contrast to other gene members which upregulated on stress, these species showed underexpression on stress (captive). Captive species showed downregulation than the natural species. Completely contrary to these findings, in Tamilnadu and Kerala, there is no significant effect observed between the natural and captive i.e., their expression levels seems to be same.
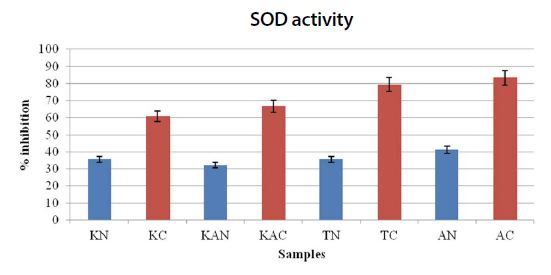
Graph showing the % inhibition of CAT activity. K: Kerala; KA: Karnataka; T: Tamil Nadu; A: Andhra Pradesh. N: Natural; C: Captive. All the values are average of triplciates. Values are expressed as value±SE.
DISCUSSION
Like most insects, honey bees also rely on detoxification enzymes like cytochrome P450s (CYPs) and carboxyl/cholinesterase (CCEs) to help themselves from toxicity caused by xenobiotic compounds in the environment. Many of these genes are studied for their functions in several insects (Claudianos et al., 2006). In this study, about 10 gene members were studied which included primary, secondary antioxidant gene members and immune related gene members. Captive bees (cultivated in apiary) are sprayed with number of miticides and xenobiotics to get rid of pests and mites. But even then these bee colonies experience colony collapse disorder reducing the overall yield in the apiaries. This is because of the strenuous stress and pressure these bees experience due to the spraying of harmful xenobiotics.
The relative concentration of the CAT members were found to be 0.16, 0.17, 0.21 and 0.13 in natural setting for Andhra, Karnataka, Tamilnadu and Kerala when compared to 0.23, 0.31, 0.45 and 0.54 in captive forms. The relative concentration of the SOD members were found to be 0.05, 0.13, 0.26 and 0.29 in natural setting for Andhra, Karnataka, Tamilnadu and Kerala when compared to 0.12, 0.19, 0.42 and 0.52 in captive forms. The main sources of ROS within living beings are aerobic metabolism, oxidative deamination of amines generated within the body, xenobiotics and their degradation within microsomes and phagocytosis. Such generated superoxide radical anions (O-2) are then converted to hydrogen peroxide (H2O2) by superoxide dismutase (SOD1/ SOD2). This hydrogen peroxide is later decomposed to water and oxygen by catalase (CAT) enzyme activity (Celic et al., 2015). Our results obtained are in accordance to the above findings in that both the CAT and SOD levels are over expressed when compared to natural setting bees. This provides us of the background to how much these little bees might have exposed to stress levels. The overexpression was too significant that cannot be neglected as well.
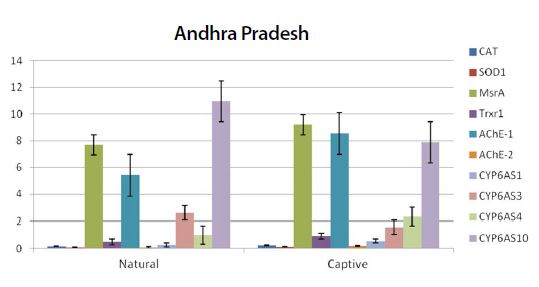
Graph showing the expression pattern of 10 gene members in the study in Natural and captive settings of Andhra Pradesh region. qPCR was performed to measure the messenger RNA expression of the gene members (n=20). The housekeeping gene RP4 was used for data normalization. Data are presented as fold change relative to the average of calibrator (P<0.05).
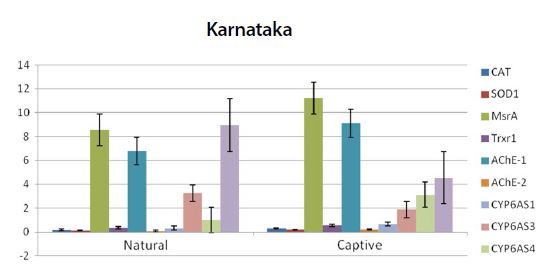
Graph showing the expression pattern of 10 gene members in the study in Natural and captive settings of Karnataka region. qPCR was performed to measure the messenger RNA expression of the gene members (n=20). The housekeeping gene RP4 was used for data normalization. Data are presented as fold change relative to the average of calibrator (P<0.05).
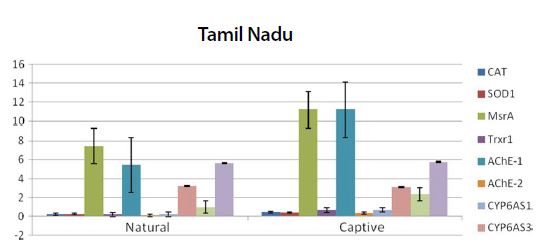
Graph showing the expression pattern of 10 gene members in the study in Natural and captive settings of Tamil Nadu region. qPCR was performed to measure the messenger RNA expression of the gene members (n=20). The housekeeping gene RP4 was used for data normalization. Data are presented as fold change relative to the average of calibrator (P<0.05).
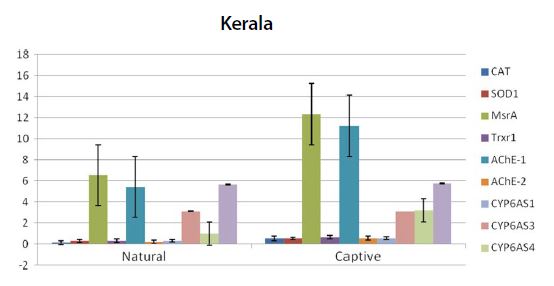
Graph showing the expression pattern of 10 gene members in the study in Natural and captive settings of Kerala region. qPCR was performed to measure the messenger RNA expression of the gene members (n=20). The housekeeping gene RP4 was used for data normalization. Data are presented as fold change relative to the average of calibrator (P<0.05).
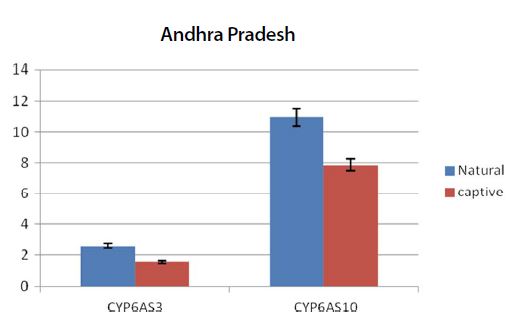
Graph showing the expression pattern of 2 gene members in the study in Natural and captive settings of Andhra pradesh. qPCR was performed to measure the messenger RNA expression of the gene members (n=20). The housekeeping gene RP4 was used for data normalization. Data are presented as fold change relative to the average of calibrator (P<0.05).
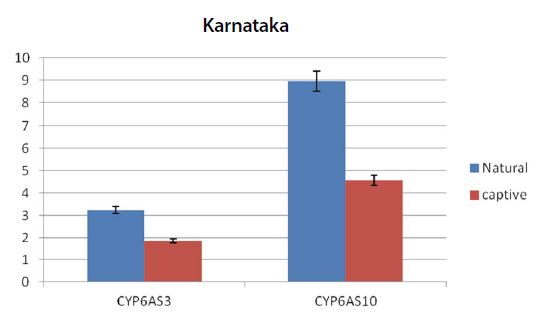
Graph showing the expression pattern of 2 gene members in the study in Natural and captive settings of Karnataka. qPCR was performed to measure the messenger RNA expression of the gene members (n=20). The housekeeping gene RP4 was used for data normalization. Data are presented as fold change relative to the average of calibrator (P<0.05).
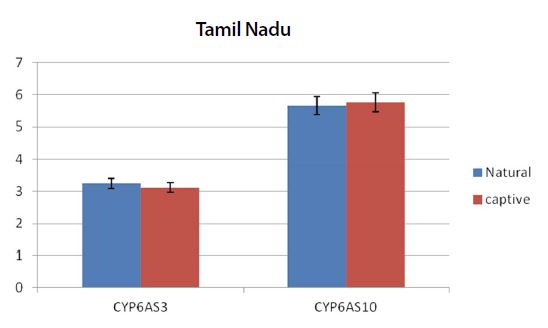
Graph showing the expression pattern of 2 gene members in the study in Natural and captive settings of Tamil Nadu. qPCR was performed to measure the messenger RNA expression of the gene members (n=20). The housekeeping gene RP4 was used for data normalization. Data are presented as fold change relative to the average of calibrator (P<0.05).
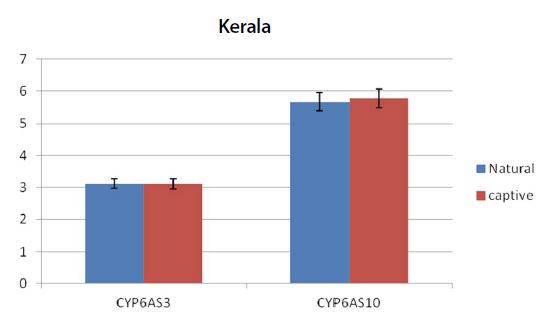
Graph showing the expression pattern of 2 gene members in the study in Natural and captive settings of kerala. qPCR was performed to measure the messenger RNA expression of the gene members (n=20). The housekeeping gene RP4 was used for data normalization. Data are presented as fold change relative to the average of calibrator (P<0.05).
Severe and acute exposure of the bees to organophosphates would eventually increase the mortality and morbidity. High dosages of imidacloprid (20 ppb) was found to significantly up-regulate the express of CAT (P<0.001), TrxR1 (P<0.05) and MsrB (P<0.001) (Gregorc, 2018). Even the secondary antioxidant gene members (MsrA & Trxr1) showed same level relative expression in accordance to those of primary antioxidant gene members (CAT & SOD). The Trxr1 gene members showed expression levels of 0.48, 0.34, 0.21 and 0.32 in natural settings of Andhra, Karnataka, Tamilnadu and Kerala regions when compared to 0.91, 0.54, 0.67 and 0.65 of captive settings. Even the same results were seen in case of expression of MsrA gene member also.
AchE1, AchE2, CYP6AS1, CYP6AS3, CYP6AS4 and CYP6AS10 are the immune related gene members reported in this paper also showed concord results as like antioxidant gene members. When AChE inhibitors were used in the treatment, expression levels of AChE was seen to be upregulated in the brain and in the gut tissues. These findings confirm of the possible role of the pesticides on the physiological effects of bees (Williamson, 2013). Organophosphates (e.g., chlorpyrifos) and the carbamates (e.g., aldicarb) act as inhibitors to acetylcholinesterase (AChE) which is said to hydrolyse acetylcholine (ACh) at the synaptic cleft (Pohanka, 2011). Such inhibition of AChE inturn leads to prolonged activation of cholinergic receptors, followed by their desensitization (Pohanka, 2011).
Likewise, in parallel to the above findings, AChE members (both 1 and 2) were upregulated 3 to 4 times among the captive forms. This might end up in a possible conclusion that the captive forms are being sprayed with high dosages of organophosphates to get rid of Varroa mites.
Cytochrome P450 monooxygenases (P450) among the bees, detoxify the phytochemicals consumed along with the honey and pollen. The most predominant and active ingredient of present in the pollen is flavonol quercetin which is seen in minute amounts within the honey (Mao et al., 2009). Along with interacting within themselves, pesticides do interfere with the ability of bees to metabolize substances of diet as like how certain medicines and combinations interfere with xenobiotic metabolism in the human diet. Among the 28 P450 genes (from the CYP3 clan), seven of them (CYP6AS17, CYP9R1, CYP6AS1, CYP9Q1, CYP9Q2, CYP9Q3, and CYP4G11) were seen to get up-regulated and down regulated by quercetin treatments. CYP6AS17 and CYP9R1 were found to over expressed and the other were underexpressed by the high-quercetin treatment (Mao et al., 2017).
Similar to the above findings, we obtained results of these cytochrome members in accordance. CYP6AS1 and CYP6AS4 gene members also showed significant results in captive forms when compared to the natural settings. But in contrast CYP6AS3 and CYP6AS10 gene members showed little or no significant effect among the natural and captive forms in case of Tamilnadu and Kerala regions. There is no such possible reason for the same, but could be the dosage of the organophosphates or miticides which are used on the current study sample. On the other hand, in case of Andhra and Karnataka the captive samples showed downregulation of the above two mentioned gene members compared to natural forms.
These findings suggest of the possible function of these gene members in regulating the stress levels in honey bees exposed to over dosages of miticides. In south India, bee keeping aviaries use over dosages of organophosphates and miticides to protecting the pollinator from the acute toxicity of not only compounds generated by Varroa mite infestation, but also miticide residues. Hence this study could end up with a possible solution of how effectively we can address this mite infestation without disturbing the bee behaviour.
CONCLUSION
The honey bee (Apis cerana L.) is a social insect which owes tremendous ecological benefit t both environment and human society. But however, these sweet little ones are experiencing a serious threat by the synergistic action of pathogens and parasites, leading to large scale colony losses, sometimes called colony collapse disorder (CCD). The Varroa mite (Varroa destructor), an ecto-parasite for these bees is the major problem in the Apiary industry, making the bee keepers to spray excess amount of miticides to protect the bees. Even though it could reduce the mites load, there is an underlying danger to the bees, where they are experiencing huge stress levels. This could lead to the metabolic functions loss and overall yield in honey. Therefore, understanding of honey bees’ ability to detoxify different pesticides is very much essential for breeding new colonies in response to the Varroa mite infestation as well as miticide management in the apiculture.
References
-
Abele, D., K. Heise, H. Pörtner and S. Puntarulo. 2002. Temperature-dependence of mitochondrial function and production of reactive oxygen species in the intertidal mud clam Mya arenaria. J. Exp. Biol. 205(13): 1831.
[https://doi.org/10.1242/jeb.205.13.1831]

-
Berenbaum, M. R. and R. M. Johnson. 2015. Xenobiotic detoxification pathways in honey bees. Cur. Opin. Insect Sci. 10: 51-58.
[https://doi.org/10.1016/j.cois.2015.03.005]

-
Celic, T., J. Purac, S. Orčič, D. Kojič, D. Vujanovic, Z. Stanimirovic, I. Gržetić, K. Ilijević, B. Sikoparija and D. Blagojević. 2015. Environmental effects on superoxide dismutase and catalase activity and expression in honey bee. Arch. Insect Biochem. Physiol. 90(4): 181-194. DOI: 10.1002/arch.21253.
[https://doi.org/10.1002/arch.21253]

-
Claudianos, C., H. Ranson, R. M. Johnson, S. Biswas, M. A. Schuler, M. R. Berenbaum, R. Feyereisen and J. G. Oakeshott. 2006. A deficit of detoxification enzymes: pesticide sensitivity and environmental response in the honeybee. Insect Mol. Biol. 15: 615-636.
[https://doi.org/10.1111/j.1365-2583.2006.00672.x]

-
Comhair, S. A. A., W. Xu, S. Ghosh, F. B. J. M. Thunnissen, A. Almasan, W. J. Calhoun, A. J. Janocha, L. Zheng, S. L. Hazen and S. C. Erzurum. 2005. Superoxide dismutase inactivation in pathophysiology of asthmatic airway remodeling and reactivity. Am. J. Pathol. 166: 663-674.
[https://doi.org/10.1016/S0002-9440(10)62288-2]

-
Douglas, T., D. S. Daniel, B. K. Parida, C. Jagannath and S. Dhandayuthapani. 2004. Methionine sulfoxide reductase A (MsrA) deficiency affects the survival of Mycobacterium smegmatis within macrophages. J. Bacteriol. 186(11): 3590-3598.
[https://doi.org/10.1128/JB.186.11.3590-3598.2004]

-
Garibaldi, L. A., I. Steffan-Dewenter, R. Winfree, M. A. Aizen, R. Bommarco, S. A. Cunningham, C. Kremen, L. G. Carvalheiro, L. D. Harder, O. Afik, I. Bartomeus, F. Benjamin, V. Boreux, D. Cariveau, N. P. Chacoff, Jan H. Dudenhöffer, B. M. Freitas, J. Ghazoul, S. Greenleaf, J. Hipólito, A. Holzschuh, B. Howlett, R. Isaacs, S. K. Javorek, C. M. Kennedy, K. M. Krewenka, S. Krishnan, Y. Mandelik, M. M. Mayfield, I. Motzke, T. Munyuli, B. A. Nault, M. Otieno, J. Petersen, G. Pisanty, S. G. Potts, R. Rader, T. H. Ricketts, M. Rundlöf, C. L. Seymour, C. Schüepp, H. Szentgyörgyi, H. Taki, T. Tscharntke, C. H. Vergara, B. F. Viana, T. C. Wanger, C. Westphal, N. Williams and A. M. Klein. 2013. Wild pollinators enhance fruit set of crops regardless of honey bee abundance. Science 339: 1608-1611. DOI: 10.1126/science.1230200.
[https://doi.org/10.1126/science.1230200]

-
Gregorc, A., M. Alburaki, N. Rinderer, B. Sampson, P. R. Knight, S. Karim and J. Adamczyk. 2018. Effects of coumaphos and imidacloprid on honey bee (Hymenoptera: Apidae) lifespan and antioxidant gene regulations in laboratory experiments. Sci. Rep. 8: 15003. DOI: 10.1038/s41598-018-33348-4.
[https://doi.org/10.1038/s41598-018-33348-4]

-
Jiang, S., T. Robertson, M. Mostajeran, A. J. Robertson and X. Qiu. 2016. Differential gene expression of two extreme honey bee (Apis mellifera) colonies showing Varroa tolerance and susceptibility. Insect Mol. Biol. 25(3): 272-282.
[https://doi.org/10.1111/imb.12217]

-
Jobin, M. P., D. Garmyn, C. Diviès and J. Guzzo. 1999. Expression of the Oenococcus oeni trxA gene is induced by hydrogen peroxide and heat shock. Microbiology 145(5): 1245-1251.
[https://doi.org/10.1099/13500872-145-5-1245]

-
Johnson, R. M., L. Dahlgren, B. D. Siegfried and M. D. Ellis. 2013. Acaricide, fungicide and drug interactions in honey bees (Apis mellifera). PLoS ONE 8(1): e54092.
[https://doi.org/10.1371/journal.pone.0054092]

-
Johnson R .M., M. D. Ellis, C. A. Mullin and M. Frazier. 2010. Pesticides and Bee Toxicity - USA. Apidologie 41: 312-331.
[https://doi.org/10.1051/apido/2010018]

-
Johnson, R. M., Z. Wen, M. A. Schuler and M. R. Berenbaum. 2006. Mediation of pyrethroid insecticide toxicity to honey bees (Hymenoptera: Apidae) by cytochrome P450 monooxygenases. J. Econ. Entomol. 99: 1046-1050.
[https://doi.org/10.1093/jee/99.4.1046]

-
Kim, S., K. Kim, J. H. Lee, S. H. Han and S. H. Lee. 2019. Differential expression of acetylcholinesterase 1 in response to various stress factors in honey bee workers. Sci. Rep. 9: 10342. DOI: 10.1038/s41598-019-46842-0.
[https://doi.org/10.1038/s41598-019-46842-0]

-
Kim, Y. H., D. H. Kwon, H. M. Ahn, Y. H. Koh and S. H. Lee.2014. Induction of soluble AChE expression via alternative splicing by chemical stress in Drosophila melanogaster. Insect Biochem. Mol. Biol. 48: 75-82. DOI: 10.1016/j.ibmb.2014.03.001.
[https://doi.org/10.1016/j.ibmb.2014.03.001]

-
Kim, Y. H., J. H. Kim, K. Kim and S. H. Lee. 2017. Expression of acetylcholinesterase 1 is associated with brood rearing status in the honey bee, Apis mellifera. Sci. Rep.-Uk 7: 39864. DOI: 10.1038/srep39864.
[https://doi.org/10.1038/srep39864]

- Li, J. H. 2015. Analysis of the factors affecting honeybee pollination efficiency for agricultural crops. J. Agric. 5(9): 104-109.
-
Li, X., W. Ma, J. Shen, D. Long, Y. Feng, W. Su, K. Xu, Y. Du and Y. Jiang. 2019. Tolerance and response of two honeybee species Apis cerana and Apis mellifera to high temperature and relative humidity. PLoS ONE 14(6): e0217921. DOI: 10.1371/journal.pone.0217921
[https://doi.org/10.1371/journal.pone.0217921]

-
Mao, W., M. A. Schuler and M. R. Berenbaum. 2017. Disruption of quercetin metabolism by fungicide affects energy production in honey bees (Apis mellifera). Proc. Natl. Acad. Sci. USA 114: 2538-2543.
[https://doi.org/10.1073/pnas.1614864114]

-
Mao, W., S. G. Rupasinghe, R. M. Johnson, A. R. Zangerl, M. A. Schuler and M. R. Berenbaum. 2009. Quercetin-metabolizing CYP6AS enzymes of the pollinator Apis mellifera (Hymenoptera: Apidae). Comp. Biochem. Physiol. B: Biochem. Mol. Biol. 154: 427-434.
[https://doi.org/10.1016/j.cbpb.2009.08.008]

- Mehran, M. J., H. Zendehbad and S. Malla. 2014. Free radical scavenging and antioxidant potential activity of cassava plants. Asian J. Pharm. Clin. Res. 7(1): 66-70.
-
Ollerton, J., V. Price, W. S. Armbruster, J. Memmott, S. Watts, N. M. Waser, O. Totland, D. Goulson, R. Alarcón, J. C. Stout and S. Tarrant. 2012. Overplaying the role of honey bees as pollinators: a comment on Aebi and Neumann (2011). Trends Ecol. Evol. 27: 141-142. DOI: 10.1016/j.tree.2011.12.001
[https://doi.org/10.1016/j.tree.2011.12.001]

-
Perez-Campo, R., M. Lopez-Torres, S. Cadenas, C. Rojas and G. Barja. 1998. The rate of free radical production as a determinant of the rate of aging: evidence from the comparative approach. J Comp Physiol [B] 168: 149-158.
[https://doi.org/10.1007/s003600050131]

-
Pohanka, M. 2011. Cholinesterases, a target of pharmacology and toxicology. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 155(3): 219-223.
[https://doi.org/10.5507/bp.2011.036]

-
Schöning, C., S. Gisder, S. Geiselhardt, I. Kretschmann, K. Bienefeld, M. Hilker and E. Genersch. 2012. Evidence for damage-dependent hygienic behaviour towards Varroa destructorparasitised brood in the western honey bee, Apis mellifera. J. Exp. Biol. 215: 264-271.
[https://doi.org/10.1242/jeb.062562]

-
Serrano, L. M., D. Molenaar, M. Wels and B. Teusink. 2007. Thioredoxin reductase is a key factor in the oxidative stress response of Lactobacillus plantarum WCFS1. Microb. Cell Fact. 6: 29. DOI: 10.1186/1475-2859-6-29
[https://doi.org/10.1186/1475-2859-6-29]

-
Tamburro, A., N. Allocati, M. Masulli, D. Rotilio, C. Di Ilio and B. Favaloro. 2001. Bacterial peptide methionine sulphoxide reductase: co-induction with glutathione S-transferase during chemical stress conditions. Biochem. J. 360(3): 675-681.
[https://doi.org/10.1042/bj3600675]

-
Valko, M., C. J. Rhodes, J. Moncol, M. Izakovic and M. Mazur. 2006. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 160: 1-40.
[https://doi.org/10.1016/j.cbi.2005.12.009]

-
Weirich, G. F., A. M. Collins and V. P. Williams. 2002. Antioxidant enzymes in the honey bee, Apis mellifera. Apidologie 33(1): 3-14. DOI: 10.1051/apido:2001001.
[https://doi.org/10.1051/apido:2001001]

-
Williamson, S. M., C. Moffat, M. A. Gomersall, N. Saranzewa, C. N. Connolly and G. A. Wright. 2013. Exposure to acetylcholinesterase inhibitors alters the physiology and motor function of honeybees. Front. Physiol. 4: 13. DOI: 10.3389/fphys.2013.00013.
[https://doi.org/10.3389/fphys.2013.00013]