Detection of Sacbrood Virus Infecting Apis cerana (AcSBV) Using Polyclonal Antibody Raised against Recombinant VP3 Capsid Protein Expressed in Escherichia coli
Abstract
In Korea, sacbrood virus infecting Apis cerana (AcSBV) is a small RNA virus belonging to the family Iflaviridae. AcSBV causes larval death, and even the collapse of entire bee colonies and its viral capsid is composed of four structural proteins (VP1, VP2, VP3 and VP4). In this study, we synthesized VP3 gene based on genomic sequences to insert into a His-tag expression plasmid of pET-28a. The resultant recombinant vector was then transformed into Escherichia coli BL21 (DE3) to express the recombinant VP3 protein (rVP3). The rVP3 protein was strongly expressed in an insoluble fraction, with a molecular size of around 26 kDa. Purification of the rVP3 protein using an affinity Ni-NTA resin, followed by SDS-PAGE separation, resulted in a high purity of the protein and used as an antigen to raise the polyclonal antibodies. Mice were then immunized with the purified rVP3 protein and the antiserum collected at 4th week and 6th week was able to detect the rVP3 at a concentration of 500 ng and 250 ng. Furthermore, IgG antibody levels reacted with wild-type larva crude protein extract, AcSBV-infected larva crude protein extracts and rVP3 protein were quantitatively analyzed by ELISA and qualitatively analyzed by western blotting. This is the first report on production of polyclonal antibodies directed against the rVP3 protein of AcSBV and its use for detection and diagnosis of virus from field samples.
Keywords:
Apis cerana, Sacbrood virus, VP3 protein, Polyclonal antibody, Detection, DiagnosisINTRODUCTION
Sacbrood virus (SBV) is one of the most destructive honeybee viruses that have a considerable impact on Asian apiculture (Ma et al., 2010; Ai et al., 2012; Forsgren et al., 2015; Wu et al., 2015; Yanez et al., 2015; Ha et al., 2016; Sarwar, 2016; Wang et al., 2016). Its infection can alter adult worker bees’ behavior such as accelerating progression from brood tending to foraging and aversion to collecting pollen (Rortais et al., 2006) and also cause failure to pupate and death during the larval stage (Li et al., 2016). SBV isolates infecting Apis cerana (AcSBV) and A. mellifera (AmSBV) represent two different serotypes (Roberts and Anderson, 2014). In a previous cross-infection study, AmSBV and AcSBV were pathogenic only in A. mellifera larvae and A. cerana larvae, respectively (Zhang et al., 2014). However, a recent study showed that AcSBV can infect A. mellifera with low epidemic and pathogenicity (Gong et al., 2016). Furthermore, the phylogenetic tree analyses supported the cross-infection of AmSBV and AcSBV (Li et al., 2016; Reddy et al., 2017; Aruna et al., 2018).
SBV belongs to the family Ifaviridae (Procházková et al., 2018). It has a linear, positive single-stranded RNA genome which is composed of 8,832 nucleotides with poly A tails at its 3ʹ end (Ghosh et al., 1999). The genome encodes a single open reading frame (a polyprotein of 2,858 amino acids) that is cotranslationally and posttranslationally cleaved by viral proteases into functional subunits (Ghosh et al., 1999). In China, Chinese sacbrood virus (CSBV) was first reported in Guangdong province in 1972 and reemerged in Liaoning Province in 2008, causing great losses in A. cerana apiculture (Zhang, 2011). Except for the 3ʹ and 5ʹ ends, the CSBV genome sequence was 8740 nucleotides, which contains a single, large open reading frame encoding a polyprotein of 2,861 amino acids (AF469603). The Liaoning CSBV is referred to as CSBV-LN to distinguish it from CSBV-GZ (AF469603) and SBV-UK (AF092924.1) according to the phylogenetic analysis and comparison of characteristics (Ma et al., 2011). In Korea, complete genome of AmSBV-Kor19 (JQ390592) and AmSBV-Kor21 (JQ390591) was reported (Choe et al., 2012a). In recent years, the population of A. cerana is decreasing close to extinction due to the lethal SBV disease which has caused more than 90% of mortality since the first observation in 2008 (Kim et al., 2008; Choi et al., 2010; Choe et al., 2012b; Jung and Cho, 2015; Theisen-Jones and Bienefeld, 2016). The causative virus of Korean honeybees was proposed Korean Sacbrood virus (KSBV) (Han et al., 2011; Lee et al., 2017) and the complete RNA genome was fully sequenced (Choe et al., 2012b).
Several detection methods have been devised to identify the presence of bee viruses. Since entire genome sequences of SBV was available (Grabensteiner et al., 2001), reverse transcriptase (RT)-PCR detection method has been used as the most common tool for the detection of the viral genome (Grabensteiner et al., 2001; Chen et al., 2004; Topley et al., 2005; Shen et al., 2005; Antnez et al., 2006; Bernyi et al., 2006; Grabensteiner et al., 2007; Kim et al., 2008) and real-time RT-PCR method has been used as the most reliable tool for the quantitation of SBV (Kukielka and Sanchez-Vizcaino, 2009; Nguyen et al., 2009; Kim et al., 2009; Phu et al., 2010; Yoo et al., 2012). In addition, the presence of AcSBV was detected in A. cerana by utilizing RT-PCR detection method (Choi et al., 2010). Recently, quantitative real-time PCR assay has been developed to detect a AcSBV-specific genomic region from infected larvae (Han et al., 2011). Nevertheless, these methods cannot provide rapid diagnosis for bee farmers at the time and site of an encounter with environmental larval honeybee samples. Therefore, a point-of-care (POC) test using antibody is required for the rapid virus diagnosis.
To date, no anti-AcSBV antibodies were commercially available. No immunological assays have been reported to detect AcSBV due to difficulties of purification for a large amount of virus particles to produce antisera. In fact, virus purification might be a lengthy procedure and provide occasionally unsatisfactory results concerning specificity and titer of the prepared antiserum due to the variable virus purity (Fajardo et al., 2007). Foreign gene expression in Escherichia coli is relatively simple, rapid and inexpensive for producing high quantities of proteins with minimal post-translational modifications (Hartley, 2006). In addition, fusion of target genes with tags facilitates their expression and improves their solubility, stability and purification (Butt et al., 2005).
In the N-terminal of SBV polyprotein, four structural proteins (VP1, VP2, VP3 and VP4) were predicted at its capsid (Ma et al., 2011), although the VP4 protein has not been found (Ghosh et al., 1999; Ma et al., 2011). A structural study shows that SBV virions do not have VP4 structural subunits that facilitate the genome delivery of typical picornaviruses (Procházková et al., 2018). Recently, the yeast two-hybrid method revealed that CSBV VP3 protein interacted with galectin (GAL), a protein involved in immune regulation and host-pathogen interaction (Fei et al., 2018). Therefore, we synthesized VP3 structural protein gene using genomic sequences and cloned into pET-28a E. coli expression system to produce recombinant VP3 protein (rVP3). Following its expression and purification, we produced polyclonal antibody against the rVP3 protein. The final goal was to use the polyclonal antibody for the immunological diagnosis of AcSBV in larva samples collected from field.
MATERIALS AND METHODS
Synthesis of VP3 gene
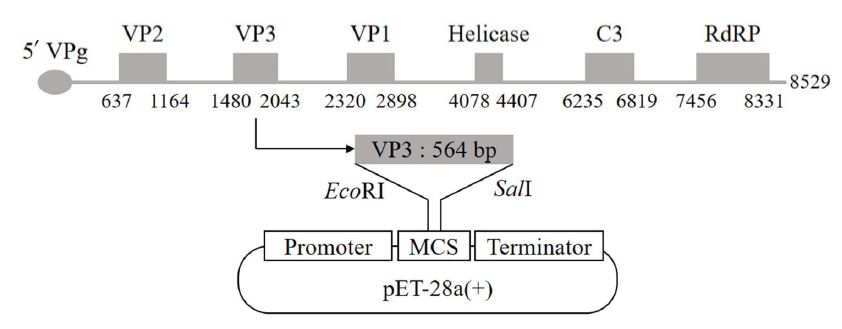
Based on the AcSBV genomic RNA sequence (HQ322114), the VP3 gene fragment (564 bp) was synthesized commercially (Bioneer Co. Daejeon, Korea), inserted into pBHA (2,002 bp, high copy vector). The recombinant cloning vector (pBHA-VP3) was transformed into E. coli strain TOP 10.
Construction of gene expression vector
To amplify the VP3 gene from the recombinant pBHA-VP3 vector, polymerase chain reaction (PCR) performed with a forward (5ʹ-TCGCGGATCCGAATTCACGTTTAATTGGGGATCAGGCGAT-3ʹ) and a reverse (5ʹ-CCGCAAGCTTGTCGACAGTTGACTGTTTTAAACCATGCAATGCAT-3ʹ) primers. The EcoRI and SalI sites (underlined) were engineered in the primers to facilitate cloning, respectively. A 50 μL PCR reaction contained 5×PCR buffer (10 μL), 10 mM dNTPs (5 μL), 10 pmol of each primer, 1 ng plasmid DNA and 0.4 μL (1 unit) SuPrime HF DNA Polymerase (GeNet Bio, Daejeon, Korea). The thermo-profile included 1 cycle of 98℃ for 2 min; 35 cycles of 98℃ for 30 s, 58℃ for 30 s, 68℃ for 2 min and additional 1 cycle of 68℃ for 5 min. The amplified VP3 DNA and pET-28a (+) (Novagen, German) were double-digested with EcoRI / SalI. Then, the VP3 DNA and linearized pET vector were purified from agarose gel and ligated to each other to construct a recombinant vector (pET28a-VP3) (Fig. 1) in frame with histidine hexamer tag. The resultant vector was transformed into E. coli BL21 (DE3) and the mixture inoculated onto Luria-Bertani (LB) agar media containing kanamycin (50 μg/mL). The selected single bacterial colonies were inoculated into 5 mL of LB medium and incubated at 37℃ for 10 h. The recombinant vector was identified by gel electrophoresis after double-digestion and PCR analyses.
Inducible expression and purification of recombinant protein
The selected single bacterial colonies were inoculated into 5 mL of LB medium and incubated at 37℃ for 10 h. The cultures (1 mL) were inoculated to 100 mL LB (50 μg/mL kanamycin) and cultured at 37℃ until the absorbance at 600 nm reached 0.6, then isopropyl β-D-1-thiogalactopyranoside (IPTG) at concentration of 1 mM was added to induce protein expression for 4 h. To increase the expression level, inducible conditions were optimized, including the concentrations of IPTG, temperatures, and induction durations. At the same time, the uninduced and vector controls were established in parallel. After induction, cells were harvested by centrifugation and the bacterial pellets were suspended in lysis buffer (10 mM imidazole, 300 mM NaCl, 50 mM NaH2PO4, pH 8.0), then lysed using 4 rounds (60 sec/each) of ultrasonication on ice (Ultrasonication power: 40 W; Probetype: Ф3 mm). After centrifugation at 8,000 rpm for 10 min, the supernatant and pellet were examined to which fraction contained the expressed protein. Both pellet and supernatant samples (20 μg) were denatured by boiling for 5 min with an equal volume of 5×sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 10% Glycerol, 1% 2-mercaptoethanol, 12.5 mM EDTA, 0.02% bromophenol blue) before separating (each sample 20 μL) on 10% SDS-PAGE. The resultant pellet was resuspended in phosphate-buffered saline (PBS; 3.2 mM Na2HPO4, 0.5 mM KH2PO4, 1.3 mM KCl, 135 mM NaCl, pH 7.4) and used for analyzing the presence of recombinant protein in inclusion bodies. AcSBV-rVP3 protein was purified from E. coli cells using Ni+-nitrilotriacetic acid (NTA) affinity column (Qiagen, USA) under denaturing conditions (7 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl, pH 8.0) as described in the manufacturer’s manual. For the final elution of 6×His-rVP3 fusion protein, we used an elution buffer E (8 M urea, 0.1 M NaH2PO4, 0.01 M Tris-HCl, pH 4.5). The efficiency of rVP3 protein extraction was analyzed by 10% SDS-PAGE of supernatants (‘cleared lysates’) after re-suspending an aliquot in 5×sample buffer as described above. The gels were viewed after staining with Coomassie Blue staining solution (0.05% Coomassie brilliant blue R-250 was dissolved in methanol, glacial acetic acid and water in 5 : 1 : 5 ratio, v/v/v) and destained in a solution of 10% glacial acetic acid and 15% methanol. Protein concentration was estimated by Bradford dye-binding assay (Bio-Rad, USA) using bovine serum albumin as a standard. The purified rVP3 was further separated by SDS-PAGE to remove the minor protein, and the targeted recombinant protein band was excised and then electro-eluted from the gel. After dialysis of rVP3 against PBS to remove SDS, allowing the protein to be used as antigen for developing the polyclonal antibody.
Production of polyclonal antibody
All animal procedures were approved by the Ethics Committee of Pai Chai University. Prior to injection, the purified rVP3 protein was dialyzed twice against 1.5 L PBS for 1.5 h at 4℃, and then used as an immunogen. Polyclonal antiserum was prepared by injecting three BALB/c mice (male, six-week-old) with the purified rVP3 protein. Before starting the injection schedule, the mice were bled to obtain the normal serum (pre-immune serum). Initially, 100 μg (50 μL) of the purified rVP3 protein emulsified with 50 μL of Freund̓s complete adjuvant (Sigma, USA) was injected subcutaneously, followed by two booster injections (50 μg/mL) with (2nd injection) and without (3rd injection) Freund’s complete adjuvant. A total of three immunizations were given to mice in 2 week intervals, and serum was recovered from the mice blood one week after every injection. Furthermore, the immunoglobulin G (IgG) was purified using Montage Antibody Purification Kit and Prosep-G media according to the manufacturer’s protocol (Millipore Co., USA). The concentration of AcSBV-rVP3-IgG was adjusted to 1 mg/mL and mixed with glycerol (50%, v/v) and stored at -20℃ until use.
Western blot analysis
The purified rVP3 proteins were separated by 10% SDS-PAGE gels. Crude protein extracts from healthy larva, AvSBV-infected larva samples (Daejeon and Samcheok, Korea) were included. The unstained gel was blotted onto a nitrocellulose membrane (NC) membrane (Atto, Japan) by a semi-dry electro-transfer apparatus (Thermo Fisher Scientific, USA). The membrane was as blocked with 5% skim milk in Tris-buffered saline (TBS; 137 mM sodium chloride, 20 mM Tris, pH 7.4) for 1 h at 37℃. The membrane was washed in TBST (0.1% Tween-20 in TBS) three times, and then was incubated with antiserum against rVP3 or rVP3-IgG (1 : 1,000 dilution, v/v) at 4℃ overnight. After washing three times with TBST, the membrane was incubated with goat anti-mouse phosphatase conjugate (1 : 7,000 dilution, v/v) at 37℃ for 1 h. Then, 66 μL of nitro blue tetrazolium (NBT) was added into 20 mL alkaline phosphatase buffer, before mixing with 33 μL of 5-bromo-4-chloro-3-indolylphosphate (NBT/BCIP) (Sigma-Aldrich, USA) to stain the membrane until the targeted protein bands reached the desired intensity.
ELISA
To test IgG purified from antiserum (2-, 4-and 6-week antiserum), the micro-titer plate was coated with the antigen solution (in coating buffer). A 50 μL aliquot from each sample (10 μg/mL of rVP3, 400 μg/mL of crude protein extracts from healthy and AvSBV-infected larva samples) was added into the coated plate. After overnight incubation at 4℃, the plate was washed with PBST (PBS containing 0.05% Tween-20) and blocked with PBST containing 1% BSA at room temperature for 2 h. After washing with PBST three times, each IgG (1 : 1,000, v/v in blocking buffer) was added into the plate which was incubated at room temperature for 2 h. After adding alkaline phosphatase conjugated goat anti-mouse IgG (1 : 5,000, v/v in blocking buffer, Sigma-Aldrich, USA), the plate was incubated at room temperature for 2 h. After washing with PBST three times, p-nitrophenyl phosphate (Sigma-Aldrich, USA), was added into the plate which was incubated at room temperature for 2 h. Absorbance value at 405 nm was measured by microplate reader (Bio-Rad, USA). A sample was considered positive, if its mean absorbance value was twice greater than of the healthy control. The statistical analysis was performed with one-way analysis of variance (ANOVA) using SPSS software (version 24.0, SPSS Inc., USA).
RESULTS AND DISCUSSION
Construction of expression vector
In Korea, the part of capsid protein gene of the AmSBV was cloned into pET-32a vector and its recombinant protein was overexpressed as 37 kDa in E. coli (Nguyen et al., 2012). In China, the structural proteins VP1, VP2 and VP3 of CSBV were successfully expressed in the pGEX prokaryotic expression system using codon optimization and their recombinant proteins were used for polyclonal antibody production (Fei et al., 2015). In this study, the VP3 gene of AcSBV (564 bp) was obtained by PCR amplification (Fig. 2A) and inserted into the pET-28a vector (Fig. 2B). The presence of VP3 DNA in the recombinant vector, pET28a-VP3 (Fig. 2C)) was confirmed by restriction digestion (Fig. 2D) and PCR analyses (Fig. 2E) and showed that the recombinant expression vector was successfully constructed.

pET-28a vector cloning for the expression of AcSBV-VP3 protein gene. (A) Amplified DNA product of VP3 gene DNA (lane 1) and (B) the restriction enzyme (EcoRI and SalI)-digested pET-28a vector (lane 1). (C) Ligation of the VP3 gene DNA into the vector, resulting in pET28a-VP3. Confirmation for the presence of the insert in the recombinant vector by restriction enzyme digestion (D) and PCR (E).
Expression and purification of the recombinant VP3 fusion protein
To examine the expression of the rVP3 protein, the E. coli transformant was cultured in 5 mL LB media and induced by IPTG (1 mM). SDS-PAGE analysis showed that the expression of the rVP3 protein was found to be abundant in the total extract and in the insoluble fraction of the pellet debris of E. coli, with a molecular size of around 26.2 kDa, but was not presented in the supernatant as a soluble protein fraction of transformant (Fig. 3A) or in the pellet of wild-type E. coli (Fig. 3B). It suggests that cytoplasmic expression is not preferred for the rVP3 protein in E. coli. To purify the rVP3, we treated urea to denature the inclusion bodies and performed a purification system using the Ni-NTA column. The rVP3 protein could be purified effectively after washing two times and elution two times (Fig. 3C).
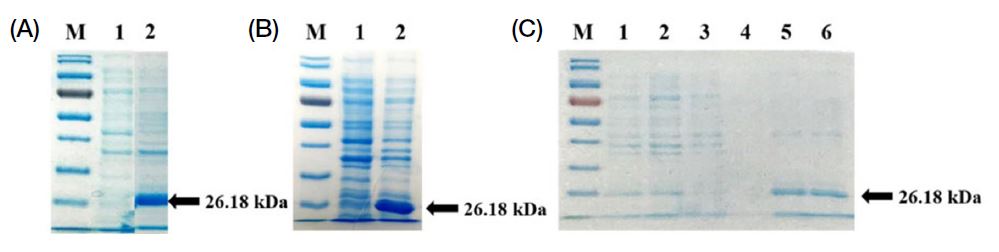
IPTG (1 mM) induction for the expression of pET-28a vector containing AcSBV-VP3 gene. (A) Soluble fraction (lane 1) and insoluble fraction (lane 2) of the Escherichia coli BL21 (DE3) transformant. (B) Insoluble fraction of wild-type (lane 1) and transformant (lane 2) E. coli BL21(DE3). (C) Purification of the recombinant VP3 (rVP3) protein by Ni-NTA resin chromatography under denaturing conditions. M: protein molecular weight marker, lanes 1-2: cell lysate 1 and 2, lanes 3-4: 1st washing and 2nd washing and lane 5-6: 1st elution and 2nd elution.
Production of the polyclonal antibody and its sensitivity and specificity
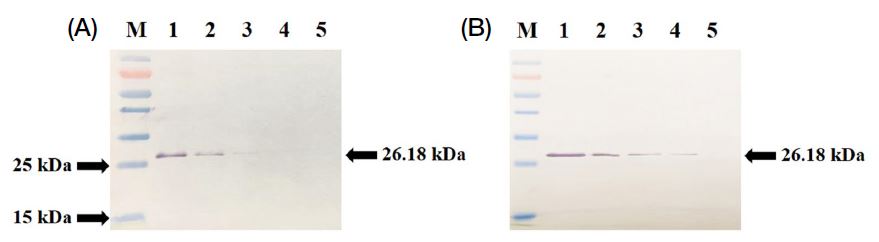
To compare the sensitivity of the raised antiserum against the rVP3, the polyclonal antiserum collected at 4th and 6th week, respectively, was used for western blot analysis. The polyclonal antiserum collected at 4th week reacted with the antigen rVP3 protein with a molecular size of 26.2 kDa at a concentration up to 500 ng (Fig. 4A, lane 3), while the polyclonal antiserum collected at 6th week reacted well with the rVP3 protein at a concentration up to 250 ng (Fig. 4B, lane 4). However, when a pre-immune antiserum was used in western blot analysis, no corresponding protein bands were observed (data not shown). These results suggest that the polyclonal antiserum against the rVP3 protein was successfully produced, making it suitable for the detection of AcSBV infection in beekeeping practices. The polyclonal antiserum was further purified to IgG to improve its sensitivity for western blot and ELISA analyses.
Efficiency test of anti-AcSBV-rVP3-IgG by ELISA and western blot
As expected, the purified rVP3 protein injection into mice effectively induced specific antibody (anti-AcSBV-rVP3 IgG) that was used as the first antibody for western blot and ELISA analyses. In the ELISA test (Fig. 5A), no significant differences were observed in PBS control, wild-type larva crude protein extract, AcSBV-infected larva crude protein extract and rVP3 protein on the IgG purified from 0 (preimmune), 2nd, 4th and 6th week sera, respectively. When compared with wild-type larva crude protein extract, the rVP3 protein showed significantly higher level of IgG purified from the 2nd week antiserum (p<0.05) and the 4th week and 6th week antisera (p<0.0001). When compared with wild-type larva crude protein extract, the infected larva crude protein extract also showed significantly higher level of IgG purified from 2nd antiserum (p<0.05) and 4th and 6th week antisera (p<0.0001). There are no statistically significant differences of IgG level between infected larva crude protein extract and rVP3 protein, although the maximum IgG level was observed in the rVP3 protein on 6th week antiserum. Similarly, the western blot results showed that the IgG purified from the 6th week antiserum reacted specifically with the rVP3 protein and AcSBV-infected larva crude protein extracts but did not react with wild-type larva crude protein extract (Fig. 5B).

(A) Abilities of anti-AcSBV-rVP3 IgG purified from the antiserum that was collected in the immunized mice with the recombinant VP3 (rVP3) protein at 2nd, 4th and 6th week, respectively. *Significantly different than the PBS control (p<0.05). ***Significantly different than the PBS control (p<0.001). (B) Western blot analysis of the rVP3 protein (5 μg, lane 1) and crude protein extract from wild-type larva (20 μg, lane 2), ACSBV-infected larva collected from Daejeon (20 μg, lanes 3~5) and Samcheok (20 μg, lanes 6~9) using the IgG purified from polyclonal antiserum collected at 6th week.
Six viruses have been identified in Korean A. cerana samples, among those Black queen cell virus (BQCV) is the most prevalent virus on adult bee samples and SBV is the most prevalent virus in larvae samples (Choe et al., 2012c). In Picornaviruses, the structural proteins play an important role in viral entry related with host cell binding (Tuthill et al., 2010), however the study of SBV protein is not sufficient, especially for structural proteins. This study provides immunological basis for developing new diagnostic kits, and further functional studies of the AcSBV structural proteins.
Acknowledgments
This work (Grants No. C0531132) was supported by project for Cooperative R&D between Industry, Academy, and Research Institute funded Korea Ministry of SMEs and Startups in 2017.
References
-
Ai, H., X. Yan, and R. Han, (2012), Occurrence and prevalence of seven bee viruses in Apis mellifera and Apis cerana apiaries in China, J. Invertebr. Pathol., 109, p160-164.
[https://doi.org/10.1016/j.jip.2011.10.006]

- Aruna, R., M. R. Srinivasan, V. Balasubramanian, and R. Selvarajan, (2018), Complete genome sequence of sacbrrod virus isolated from Asiatic honey bee Apis cerana indica in India, Virus Dis., 28, p453-460.
-
Butt, T. R., S. C. Edavettal, J. P. Hall, and M. R. Mattern, (2005), SUMO fusion technology for difficult-to-express proteins, Protein Expre. Purif., 43, p1-9.
[https://doi.org/10.1016/j.pep.2005.03.016]

-
Chen, Y., Y. Zhao, J. Hammond, H. T. Hsu, J. Evans, and M. Feldlaufer, (2004), Multiple virus infections in the honey bee and genome divergence of honey bee viruses, J. Invertebr. Pathol., 87, p84-93.
[https://doi.org/10.1016/j.jip.2004.07.005]

- Choe, S. E., L. T. Nguyen, J. H. Noh, C. H. Kweon, K. E. Reddy, H. B. Koh, K. Y. Chang, and S. W. Kang, (2012a), Analysis of the complete genome sequence of two Korean Sacbrood viruses in the honey bee, Apis mellifera, Virology, 432, p155-161.
- Choe, S. E., T. T. Nguyen, B. H. Hyun, J. H. Noh, H. S. Lee, C. H. Lee, and S. W. Kang, (2012b), Genetic and phylogenetic analysis of South Korean sacbrood virus isolates from infected honey bees (Apis cerana), Vet. Microbiol., 157, p32-40.
- Choe, S. E., L. T. Nguyen, J. H. Noh, H. B. Koh, Y. H. Jean, C. H. Kweon, and S. W. Kang, (2012c), Prevalence and distribution of six bee viruses in Korean Apis cerana populations, J. Invertebr. Pathol., 109, p330-333.
- Choi, Y. S., M. Y. Lee, I. P. Hong, N. S. Kim, H. K. Kim, K.G. Lee, and M. L. Lee, (2010), Occurrence of sacbrood virus in Korean apiaries from Apis cerana (Hymenoptera: Apidae), Kor. J. Apicul., 25, p187-191.
-
Fajardo, T. V. M., D. R. Barros, O. Nickel, G. B. Kuhn, and M. Zerbini, (2007), Expression of Grapevine leaf roll-associated virus 3 coat protein gene in Escherichia coli and production of polyclonal antibodies, Fitopatol. Bras., 32, p496-500.
[https://doi.org/10.1590/s0100-41582007000600007]

- Fei, D., H. Zhang, Q. Diao, L. Jiang, Q. Wang, Y. Zhong, Z. Fan, and M. Ma, (2015), Codon optimization, expression in Escherichia coli, and immunogenicity of recombinant Chinese sacbrrod virus (CSBV) structural proteins VP1, VP2, VP3, PLos ONE, 10, pe0128486.
-
Fei, D., D. Wei, X. Yu, J. Yue, M. Li, L. Sun, L. Jiang, Y. Li, Q. Diao, and M. Ma, (2018), Screening of binding proteins that interact with Chinese sacbrood virus VP3 capsid protein in Aphis cerna larvae cDNA library by the yeast two-hybrid method, Virus Res., 248, p24-30.
[https://doi.org/10.1016/j.virusres.2018.02.006]

-
Forsgren, E., S. Wei, D. Guiling, L. Zhiguang, T. V. Tran, P. T. Tang, T. A. Troung, T. Q. Dinh, and I. Fries, (2015), Preliminary observation on possible pathogen spill-over from Apis mellifera to Apis cerana, Apidologie, 46, p265-275.
[https://doi.org/10.1007/s13592-014-0320-3]

-
Ghosh, R. C., B. V. Ball, M. M. Willcocks, and M. J. Carter, (1999), The nucleotide sequence of sacbrood virus of the honeybee: an insect picorna-like virus, J. Gen.Virol., 80, p1541-1549.
[https://doi.org/10.1099/0022-1317-80-6-1541]

- Ha, T. T., T. K. L. Nguyen, T. L. Mai, H. L. Thanh, T. H. Nguyen, H. T. Pharm, E. R. Kondreddy, M. S. Yoo, Y. H. Kim, Y. S. Cho, S. W. Kang, and V. Q. Dong, (2016), Prevalence of bee viruses among Apis cerana population in Vietnam, J. Apic. Res., 55, p379-385.
- Han, S. H., Y. S. Choi, and M. L. Lee, (2011), Development of highly specific quantative real-time PCR method for the detection of Sacbrood virus in Korean honeybees, Apis cerana, J. Apicul., 26, p233-240.
-
Hartley, J. L., (2006), Cloning technologies for protein expression and purification, Curr. Opin. Biotechnol., 17, p359-366.
[https://doi.org/10.1016/j.copbio.2006.06.011]

- Jung, C., and S. K. Cho, (2015), Relationship between honeybee population and honey production in Korea, J. Apic., 30, p7-12.
- Kim, H. K., Y. S. Choi, M. L. Lee, M. Y. Lee, K. G. Lee, and N. H. Ahn, (2008), Detection of sacbrood virus (SBV) from the honeybee in Korea, Korean J. Apic., 23, p103-109.
- Kukielka, D., and J. M. Sanchez-Vizcaino, (2009), One-step real-time quantitative PCR for the detection and field study of Sacbrood honeybee and Acute bee paralysis viruses, J. Virol. Methods, 161, p240-246.
- Li, Y., Z. J. Zeng, and Z. L. Wang, (2016), Phylogenetic analysis of the honey bee scabrood virus, J. Apic. Sci., 60, p31-38.
- Ma, M., M. Li, J. Cheng, S. Yang, S. Wang, and P. Li, (2011), Molecular and biological characterization of Chinese sacbrood virus LN isolate, Comp. Funct. Genomics, 409386.
- Nguyen, P. V., B. R. Lee, and B. S. Yoon, (2012), Over-expression and purification of capsid protein of sacbrood virus in E. coli, J. Apic., 27, p209-214.
- Nguyen, T. K. C., M. S. Yoo, M. H. Kang, S. H. Han, C. H. Yun, and B. S. Yoon, (2009), Development of real time PCR assay for the detection of Sacbrood virus in honeybee (Apis mellifera L.), Korean J. Apic., 24, p15-21.
- Procházková, M., T. Fuzik, K. Škubník, J. Moravcová, Z. Ubiparip, A. Přidal, and P. Plevka, (2018), Virion structure and genome delivery mechanism, of sacbrood honeybee virus, PNAS., 30, p7759-7764.
-
Reddy, K. E., M. S. Yoo, Y. H. Kim, N. H. Kim, M. Ramya, H. N. Jung, T. B. Thao le, H. S. Lee, and S. W. Kang, (2016), Homology differences between complete Sacbrood virus genomes from infected Apis mellifera and Apis cerana honeybees in Korea, Virus Genes., 52, p281-289.
[https://doi.org/10.1007/s11262-015-1268-8]

-
Roberts, J. M. K., and D. L. Anderson, (2014), A novel strain of sacbrood virus of interest to world apiculture, J. Invertebr. Pathol., 118, p71-74.
[https://doi.org/10.1016/j.jip.2014.03.001]

- Rortais, A, D. Tentcheva, A. Papachristoforou, L. Gauthier, G. Arnold, M. Colin, and M. Bergoin, (2006), Deformed wing virus is not related to honeybees’ aggressiveness, Virol. J., 3, p61.
- Sarwar, M., (2016), Prevalence of multiple viral diseases associated with honey bees colony collapse and control of disorders, Int. J. Zool. Stud., 1, p29-34.
-
Theisen-Jones, H., and K. Bienefeld, (2016), The Asian honey bee (Apis cerana) is significantly in decline, Bee World, 93, p90-97.
[https://doi.org/10.1080/0005772x.2017.1284973]

- Tuthill, T. J., E. Groppelli, J. M. Hogle, and D. J. Rowlands, (2010), Picornaviruses. Curr. Top Microbial, Immunol., 343, p43-89.
- Wang, M., J. Bi, L. Wang, D. Zhou, X. Ma, W. Li, W. Zhao, G. Yin, J. Liu, and S. He, (2016), Prevalence of four common bee RNA viruses in Eastern bee populations in Yunnan province. China., Vet. Sci. Technol., 7, p1.
-
Wu, Y. Y., H. R. Jia, Q. Wang, P. L. Dai, Q. Y. Diao, S. F. Xu, X. Wang, and T. Zhou, (2015), Multiple virus infections and the characteristics of chronic bee paralysis virus in diseased honey bees (Apis mellifera L.) in China, J. Apic. Sci., 59, p95-106.
[https://doi.org/10.1515/jas-2015-0026]

- Yanez, O., H. Q. Zheng, X. L. Su, F. L. Hu, P. Neumann, and V. Dietermann, (2015), Potential for virus transfer betwween the honey bees Apis mellifera and A. cerana, J. Apic. Res., 54, p179-191.
-
Yoo, M. S., K. C. Thi, P. Van Nguyen, S. H. Han, S. H. Kwon, and B. S. Yoon, (2012), Rapid detection of sacbrood virus in honeybee using ultra-rapid real-time polymerase chain reaction, J. Virol. Methods, 179, p195-200.
[https://doi.org/10.1016/j.jviromet.2011.10.014]

- Zhang, X., (2011), Managed honeybee colonies and honey production in China grew during the last five decades, J. Apicul. Sci., 55, p77-85.
-
Zhang, Y, G. Zhang, X. Huang, and R. Han, (2014), Proteomic analysis of Apis cerana and Apis mellifera larvae fed with heterospecific royal jelly and by CSBV challenge, PLoS One, 9, pe102663.
[https://doi.org/10.1371/journal.pone.0102663]