Isolation of Soil Actinobacterial Strains Showing Antifungal Activity against Aspergillus flavus, a Causative Agent of Stonebrood Disease in Honeybee
Abstract
Stonebrood is a well known fungal disease of honeybee. Aspergillus flavus is the most common agent of stonebrood. In this study, we isolated more than 2,000 actinobacterial strains and screened them for antifungal activity against A. flavus by inhibition zone measurement. As a result, we found that 4 Streptomyces spp. and 1 Cupriavidus sp. showed relatively high antifungal activities with the diameters of up to 21 mm inhibition zone. Therefore, this study may play a role to control stonebrood of honeybee in the near future.
Keywords:
Stonebrood, Aspergillus flavus, Actinobacteria, Streptomyces, Cupriavidus, Antifungal activityINTRODUCTION
Honeybee is a socially important insect to participate 25% of pollination as a major pollinator for a variety of crops (Evans and Hung, 2000; Morse and Calderone, 2000). Because honeybees tend to make a group to live together with around several ten thousand individuals in a small space, they can provide a proper inhabiting environment for many microorganisms that cause high possibility of various diseases (Morse, 1978).
Chalkbrood disease and stonebrood disease are the most well-known fungal diseases (Lee et al., 2004). Stonebrood, first reported by Maassen (1916) in German, is a fungal disease occurred in Apis millifera in the state of lava or adult by infection of Aspergillus spp., mainly developed at the brood stage (Tanada and Kaya, 1993). This disease is most often infected by Aspergillus flavus among Aspergillus species (Gilliam and Vandenberg, 1990), and the color of dead larval body is yellowish green due to the spores of A. flavus. If they are infected by A. fumigatus and/or A. nidulans, the green spores can be found in the dead larval body. Since stonebrood has a similar symptom as in chalkbrood, general beekeepers have been recognized as the same disease. However, they can distinguish the two diseases through different body colors, yellowish greengreen in stonebrood versus white/black in chalkbrood. Infection occurs by entering of the spores into the digestive gut, which are germinated to be the hyphae. Then the hypae penetrate into the tissues, grow and kill the larvae that can be discovered as hard stones in shape (Lee et al., 2004).
Stonebrood disease has been accepted in general beekeepers as a disease that gives less damage than bacterial diseases, but it is reported that stonebrood disease is known as the most serious honeybee disease damaging widely in beekeeping farmers of Gangwon-do province in Korea (Yoon, 2001). In addition, in many cases, the infected larvae with stonebrood disease are very high risky in mixed infections with Jose horsemen chalk disease and viral diseases, which cause more serious problems (Yoon, 2001), and in chronic damage for many years within the hive.
Actinobacteria are Gram-positive, chain-forming, and widely distributed in soil and share common characteristics partially with bacteria and fungi. So they vary in morphological and physiological properties, with which actinobacteria produce various secondary metabolites useful as antibiotics, and greatly influenced by culture conditions and the composition of media (Wiens et al., 2009). In addition, actinobacteria not only contribute in matter cycle of ecosystems through the degradation of a wide range of substrates by enzymes, enzyme-inhibitors, immunoregulatory molecules, etc. (Jones, 1985; Beppu and Horinouchi, 1991; Shin, 1991), but they also are very valuable in a research field for industrial applications (Vacelet, 1975). Out of around 10,000 antibiotics found from microbial metabolites, actinobacteria produces over 75%, among which a genus Streptomyces occupies 74% (Datta et al., 2000).
In this study, we tried to isolate actinobacterial strains from various soil samples and tested their antifungal activity against Aspergillus flavus, an agent of stonebrood disease.
MATERIALS AND METHODS
Sample collection
Various soil samples were collected from all over the country, and the collected samples were freshly maintained in the plastic bags during transfer and kept them in the refrigerator at 4°C until use (Lee and Kim, 2014).
Isolation of actinobacteria
Across the country, we collected soil samples for finding actinobacterial strains living in soil and for extracting the nutrients to be used as soil extract. The small particles were removed through sieving, and then the suitable soils were placed in each well in a 6-transwell plate with addition of sterilized distilled water for moisturizing. After the membrane filter inserts were placed on soil layer in each well, the five liquid media were poured into each insert, respectively. These media were humic acid-vitamin medium, brain heart infusion (BHI) medium, R2A medium, Bennett’s medium and Benedict’s modification of the Lindenbein medium (Lee and Kim, 2014).
Certain supernatant from soil-mixed solution including microbes was used as an inoculum. Cultivation was done in a shaking incubator at 120rpm and 28°C for 2 weeks with refill of the liquid media periodically to avoid evaporative loss. A serial dilution was made like 10-1, 10-2, 10-3, 10-4, 10-5 and 10-6. 100μL of dilution was used for spreading on the agar plate. After incubation at 28°C for 5 days, all the different types of colonies were picked up, streaked individually on each agar plate and repeated until getting pure colonies. All the obtained pure colonies were conserved in 20% glycerol stocks at -20°C (Nguyen et al., 2013).
Test Strains
The test strain, A. flavus, was isolated from stonebrood disease-infected bees obtained from Korea Honeybee Disease Institute of Kyonggi University. To isolate A. flavus, the infected honeybees were grinded, the grinded material was spread on PDA (potato dextrose agar) plates, and the colonies were checked everyday for 5 days. The isolate was further cultivated on SDA-Y agar plate for 5 days, and then onto SDA-Y + R2A (1:1) agar plate for the inhibition zone test.
Measurement of inhibition zone
A. flavus was inoculated in SDA-Y liquid medium and cultivated in a shaking incubator for 1 day. After the measurement of OD value, the spreading on SDA-Y + R2A (1:1) agar plate was performed. Then 4 different actinobacterial isolates were inoculated with four spots on the fungal lawn plate, respectively. After the incubation for 3~5 days at 25°C, each inhibition zone (diameter) was measured by using a ruler.
Identification of microbes
Genomic DNA was extracted using an InstaGene Matrix kit (Bio-Rad, Hercules, CA, USA), and the 16S rRNA gene was amplified by PCR using the universal primer set for bacteria: 27F (5´-AGA GTT TGA TCM TGG CTC AG-3´) and 1492R (5´-TAC GGY TAC CTT GTT ACG ACT T-3´). The PCR product was purified with a multiscreen filter plate (Millipore Corp., Bedford, MA, USA) and the sequencing reaction was prepared with the PRISM BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA, USA). The reaction was incubated at 95°C for 5 min, cooled on ice for 5 min, and then analyzed with an ABI Prism 3730XL DNA Analyzer automated DNA sequencing system (Applied Biosystems). The near-complete sequence of the 16S rRNA gene was identified using the EzTaxon server (Kim et al., 2012).
To identify A. flavus, a primer set was used: Asp 18S-F (5´-ATC GGG CGG TGT TTC TAT G-3´) and Asp 18SR (5´-ACC GGG CTA TTT AAG GGC CG-3´) to get 312 bp PCR products. Then the PCR products were checked through electrophoresis (1×TAE, 1.5% agarose gel).
RESULTS AND DISCUSSION
Identification of microorganisms
Aspergillus flavus was identified by checking the specific gene size through PCR reactions with the primers and electrophoresis. Among the tested three isolates, the lane 1 was matched with the size of the gene was the same size as the positive control (lane P: A. flavus), showing 300 bp (Fig. 1).
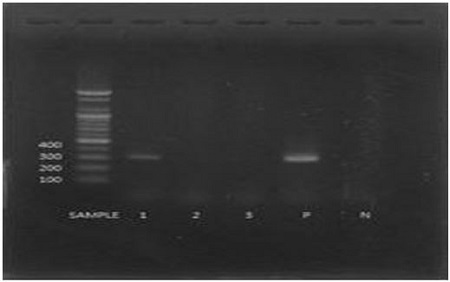
PCR test result to identify A. flavus on electrophoresis gel (lane 1~3: isolates from the larvae with stonebrood; P: positive control; N: negative control).
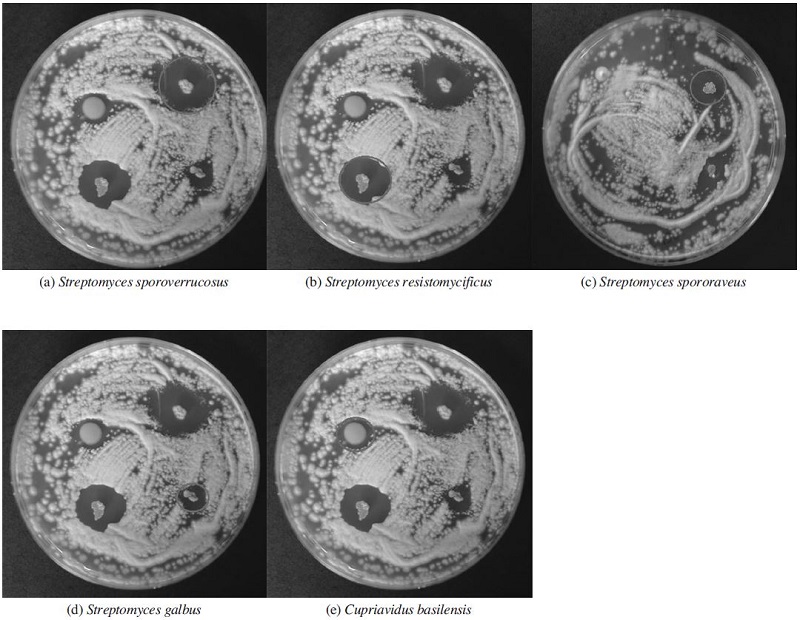
Five isolated strains having antifungal activity were identified as four Streptomyces spp. and one Cupriavidus sp. as shown in Table 1 by using 16S rRNA gene through EzTaxon. In particullar, Cupriavidus sp. L-1 can be a potential new species (98.25% similarity in 16S rRNA gene sequence) that has the antifungal activity as reported for the first time in this study.
The stonebrood of honeybee causes larval death by the toxin produced from A. flavus which can fill the body fully with hyphal growth after the spore germination in the gut. The hyphae extended from the inside of the body form the conidiospores that have white color in the beginning and then different colors according to different species: yellowish green by A. flavus, and green by A. fumigatus and A. nidulans (Samson et al., 2000).
Finding of actinbacterial strains having antifungal activity
Up to date, 80% of the antibiotics has been originated from the genus Streptomyces (Watve et al., 2001). In these days, however, the number of new antibiotics to be founded are not many.
With the goal of finding new antibiotics/compounds from antiobacteria, we have used three methods for the selection of bacteria from soil samples as mentioned in the precious research (Nguyen et al., 2013). This study constructed an actinobacterial library including around 2,000 isolates.
Some of actinobacterial isolates in the library were able to inhibit the growth of A. flavus, an agent of stonebrood disease. They showed the inhibition zones on A. flavus lawn (Fig. 2), which were measured in diameters to compare their antifungal activities against A. flavus (Table 1). These isolates are most related with Streptomycessporoverrucosus, S. resistomycificus, S. spororaveus, S. galbus, and Cupriavidusbasilensis, respectively. Among them, S. sp. T-64, S. sp. A-64 and S. sp. T-263, showed the most effective antifungal activities (Fig. 2 and Table 1). Although Cupriavidus sp. L-1 seemed to have enough size of diameter, it was excluded from the most effective strains because the size of spot was too large to consider as inhibitory area (Fig. 2). However, it is a meaningful isolate since it is a potential new species which may produce a new com-pound to control many antibiotic-resistant infectious pathogens.
Therefore, we expect that stonebrood disease may be controlled by using these actinobacterial cultures or secondary metabolites purified in the near future. Further, we may develop a new drug to control some drug-resistant microbial pathogens.
Acknowledgments
This study was supported by Bio-industry Technology Development Program (312027-3), Ministry of Agriculture, Food and Rural Affairs, by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science, and Technology (2011-0010144), and also by Kyonggi University Research Assistant Fellowship 2015.
LITERATURE CITED
-
Beppu, T., and S. Horinouchi, (1991), Molecular mechanisms in Streptomyces, Planta Medica, 57, p44-47.
[https://doi.org/10.1055/s-2006-960228]

- Datta, K., S. Shiha, and P. Chattopadhyay, (2000), Reactive oxygen species in health and disease, Natl. Med. J. India, 13, p304-310.
-
Evans, J.D., and A.C. Hung, (2000), Molecular phylogenetics and the classification of honey bee viruses, Arch. Virol, 145, p2015-2026.
[https://doi.org/10.1007/s007050070037]

- Gilliam, M., and Vandenberg, J., (1990), "Fungi", ln Morse, RA., and Nowogrodzki, R. (Eds), Honeybee pests, predators, and Disease, Second Edition, Cornell University Ress, Ithaca and London, p64-78.
- Jones, G.H., (1985), Regulation of phenoxazinone synthase expression in Streptomyces antibiotics, J. Bacteriol, 163, p1215-1221.
-
Kim, O.S., Y.J. Cho, K. Lee, S.H. Yoon, M. Kim, H. Na, S.C. Park, Y.S. Jeon, J.H. Lee, H. Yi, S. Won, and J. Chun, (2012), Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species, Int. J. Syst. Evol. Microbiol, 62, p716-721.
[https://doi.org/10.1099/ijs.0.038075-0]

- Lee, H.M., J.S. Ha, Y.H. Jo, S.H. Nam, and B.S. Yoon, (2004), PCR Detection Method of Ascosphera apis, Aspergillus flavus for Rapid Identification of Fungal Disease in Honeybee, Korean J. Apic, 19(2), p139-148.
- Lee, H.J., and J.S. Kim, (2014), Control of Chalkbrood by using Actinobacterial Culture, J. Apic, 29(1), p21-26.
- Maassen, A., (1913), Weitere Mitteilungen uber der seuchenhaften Brut-krankheiten der Bienen [Further communication on the epidemic brood disease of bees], Mitteilungen aus der Kaiserlichen Biologischen Anstalt fur Landund Forstwirtscshaft, 14, p48-58.
- Morse, R.A., (1978), Introduction, p13-22, In Honey bee pests, predators, and diseases, ed. by R. A. Morse, p430Cornell Univ, Ithaca.
- Morse, R.A, and N.W. Calderone, (2000), The value of honey bee pollination in the United States, Bee Culture, 128, p1-15.
- Nguyen, T.M., H. Lee, and J. Kim, (2013), Selective isolation of actinobacteria showing antibacterial activity against PaeniBacillus larvae from soil samples collected in South Korea, J. Apic, 266, p50-55.
- Samson, R.A., E.S. Hofkstra, J.C. Frisvad, and O. Filtenbrog, (2000), Introdution to food and airborne fungi, An institute of the Royal Netherlands Academy of Arts and Sciences, p65-73.
- Shin, J., (1991), Mid-and Long-term Reserch Plan on Marine Natural Products, KORDI.
- Tanada, Y., and H. K. Kaya, (1993), Insect Pathology, Academic press, p363.
- Vacelet, J., (1975), Etude en microscopie Electronique de l'association entre bacteries et spongiaires du genre Verongia, J. Microsc. Biol. Cell, 23, p271-288.
-
Watve, M.G., R. Tickoo, M.M. Jog, and B.D. Bhole, (2001), How many antibiotics are produced by the genus Streptomyces, Arch. Microbiol, 176, p386-390.
[https://doi.org/10.1007/s002030100345]

-
Wiens, M., P. Wrede, V.A. Grebenjuk, O.V. Kaluzhnaya, S.I. Belikov, H.C. Schroder, and W.E. Muller, (2009), Towards a molecular systematics of the Lake Baikal/ Lake Tuva sponges, Prog. Mol. Subcell. Biol, 47, p111-144.
[https://doi.org/10.1007/978-3-540-88552-8_5]

- Yoon, B., (2001), Effective for the control of bee diseases and disease prevention into national, The Korea beekeeping Bulletin, p14-16.