
Over-expression and Purification of Recombinant 3C-like Protease from Black Queen Cell Virus in Escherichia coli
Abstract
Black queen cell virus is one of the members belonging to Picornavirus, which possesses a 3C-like protease. Viral replication and capsid assembly in the order Picornavirales require polyprotein proteolysis processing, which is performed by 3C or 3C-like (3CL) proteases. Therefore, viral proteases are attractive targets for anti-viral therapy. The complete 3C-like protease gene from Black queen cell virus was cloned into pGEM-3Zf(+) vector, and the amino acid sequence revealed a high homology (99.1%) to 3C-like protease deposited in a reference Genbank EF517515.1. For the expression, 3C-like protease gene was subcloned into pMAL-C2 vector system. The expression level of MBP-BQCV-3CL protein was evaluated under different standards of cell optical density, IPTG concentration, induction time, and induction temperature. As the result, the optimal condition was confirmed as the induction with 0.1mM IPTG at 25°C for 6 hours, and a purification step using amylose resin reached a high protein yield of approximately 2.22mg per 1ml of cultured cells. Purified MBP-BQCV-3CL proteins can provide an important antigen source to generate monoclonal antibody and will become a potential candidate for anti-viral drugs, which can inactivate 3C-like protease function as well as BQCV replication.
Keywords:
Black queen cell virus (BQCV), 3C-like protease (3CL), Maltose binding protein (MBP), HoneybeeINTRODUCTION
The order Picornavirales includes five families of positive single stranded RNA viruses (Dicistroviridae, Picornaviridae, Marnaviridae, Secoviridae, and Iflaviridae) that share the similar properties.
Capsid proteins organized in a module contain three related jelly-roll domains, and non-structural proteins contain conservative three domains helicase, a cysteine proteinase with a chymotrypsin-like fold, and an RNA-dependent RNA polymerase (Le Gall et al., 2008).
Black queen cell virus belongs to the new genus Cripavirus (Family Dicistroviridae) (MayO, 2002). It was detected from queen prepupae and pupae, by the darkened areas on the wall of infected cells (Bailey and Woods, 1977). Genome organization of BQCV was reported as 8550 nucleotides polyadenylated comprising two large ORFs. The 5’-proximal ORF 1 represents 4968 nucleotides corresponding to a non-structural polyprotein, while the 3’-proximal ORF 2 encodes a capsid polyprotein (Fig. 1a). The amino acid sequences in both ORFs were reported with a high similarity to non-structural and structural proteins, respectively of Drosophila C virus (DCV), Rhopalosiphum padi virus (RhPV), Himetobi P virus (HiPV), and Plautia stali intestine virus (PSIV) (Leat et al., 2000).
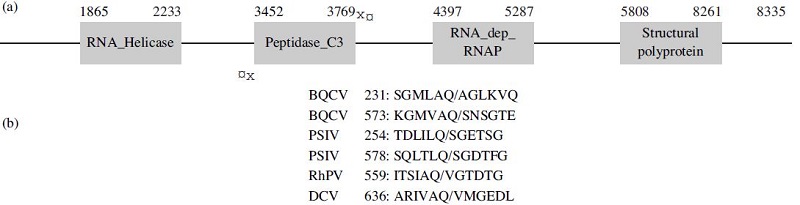
Schematic presentation of Black queen cell virus genome (Accession No. EF517515.1) and the comparison of peptide sequences of 3C-like protease cleavage sites between Honeybee viruses. (a) Genome of Black queen cell virus comprises of a non-structural polyprotein at 5’- proximal and a structural polyprotein at 3’-proximal. The numbers at the top of each box presented nucleotide sites of coding gene regions. 3C-like protease containing 318 nucleotides located between RNA_helicase and RNA dependent RNA polymerase genes. The arrows implied a pair of primer which was used for amplification of 3C-like protease gene in this study. (b) Alignment of peptide sequences at cleavage sites. The position of each cleavage site relative to the corresponding capsid polyprotein sequence is shown at the beginning of each sequence. The scissile bond of the cleavage site is presented by a forward slash. The amino acids that were emphasized by yellow colors presented the conserved sequences in cleavage sites from mentioned viruses. In particularly, amino acid glutamine (Q) existed in all sequences at the adjacent cleavage sites of each digestion.
In viral replication as well as capsid assembly, proteolysis processing plays a crucial role of catalyzing the hydrolysis of peptide bonds. After translation, the viral precursor proteins are required to be cleaved immediately to generate mature functional viral proteins (Doughterty and Semler, 1993). This process would be conducted by several proteases and a 3C or 3C-like (3CL) protease (Le Gall et al., 2008; Palmenberg, 1990).
To date, there have been many studies on the structural and functional 3C and 3C-like protease in several Picornaviruses consisting of Enterovirus A71 (Lin et al., 2012), Iflavirus (Shan et al., 2012), and Food and Mouth disease virus (Curry et al., 2007). Most of the studies have evidently found that 3C or 3C-like protease are cysteine proteases and contain chymotrypsin (Allaire et al., 1994; Cui et al., 2011; Gorbalenya et al., 1986; Malcolm, 1995). Therefore, 3C and 3C-like protease are called “chymotrypsin-like cysteine protease”.
However, among the studies on the order Picornavirales, none has concentrated on the expression and purification of 3C-like protease from Black queen cell virus. Thus, our work will be the first to show the preparation of a purified 3C-like protease protein, and it will serve as an important material for further studies.
MATERIALS AND METHODS
Collection of Honeybee samples
Adult Honeybees A. mellifera infected by BQCV were collected from various South Korean apiaries in 2013. These samples were selected by an identification step using PCR and were stored in freezer at -70°C until processed.
RNA extraction and Reverse transcription
Less than 3 bees were homogenized by MagNA Lyser green Beads (Roche, Switzerland) and total RNA were extracted by using AllspinTM for total DNA & RNA isolation from tissues and cultured cells (GeneAll, Korea), according to the manufacturer’s instructions. Total RNAs concentration was determined by Biophotometer (Eppendorf, Germany).
The extracted RNAs were reverse transcribed by AccuPower£ RT Premix (Bioneer, Korea). Briefly, 2μg of total RNAs were incubated with 200 pmole of OligodT (Bionics, Korea) at 70°C for 5 min, and then the cDNA synthesis reaction was performed at 42°C for 60 min and at 94°C for 5 min. Subsequently, cDNAs were ready for experiments.
Construction of 3C-like protease (3CL)gene from BQCV in pGEM-3Zf(+) vector
A full-length (348bp) gene encoding BQCV-3CL protease was amplified by PCR using BQCV cDNA. This sequence included an intergenic of 30 nucleotides. The forward and reverse primers for BQCV-3CL protease PCR containing BamHI and SalI restriction enzymes were designed, based on a deposited GenBank Accession No. EF517515.1 . Details about the primers are shown in Table 1.
The conditions for PCR were as follows: a denaturation at 94°C for 5 minute, 35 cycles at 94°C for 30 second, 55°C for 30 second, 72°C for 30 second, and final postextension at 72°C for 10 minute.
The amplified BQCV-3CL protease fragments were cloned into pGEM-3Zf(+) vector (Promega, USA). The recombinant clones were transformed to competent cells DH5α. The clones were examined by a colony PCR and restriction analysis. Finally, the sequence of recombinant BQCV-3CL protease clones were verified by DNAsequencing.
Expression of MBP-BQCV-3CL protein
For the expression, the BQCV-3CL protease genes were transferred into pMAL-C2 expression vector (New England Biolabs, USA). E. coli BL21 (DE3) pLysS with recombinant MBP-BQCV-3CL protease genes were cultured in LB medium including Ampicillin (0.05mg/ml) and Chloramphenicol (0.05mg/ml) at 37°C for overnight. A bacterial stock was prepared after overnight culture, and then 1ml of bacterial culture was immediately subcultured into 25ml of LB medium. The cells were induced by 0.6mM IPTG at 25°C for 6 hours, until the optical density of cell (OD600) reached 0.6. The cells were collected by a centrifugation at 4°C with 3000 rpm for 15 minutes and were suspended in 1X PBS buffer. After sonication, proteins from supernatant were stained with 5X probe buffer, denaturated at 99°C for 10 minutes, and then loaded onto SDS-PAGE.
To optimize the conditions for BQCV-3CL protease expression, bacterial cultures were grown with different optical densities at 600nm (OD600) and were induced with different concentrations of IPTG as follows: 0.1, 0.3, 0.6, 1, and 2mM. Cells were collected at 1, 3, 6, 12, and 24 hours after inducing at 25°C. The induction temperature was evaluated at 25, 30, and 37°C.
Purification of MBP-BQCV-3CL protein
The fusion MBP-BQCV-3CL proteins were purified by an amylose resin (Elpis, Korea) under an optimal expression condition. The IPTG induced cells were harvested by a centrifugation, and the bacterial pellets were suspended in PBS buffer. The cell supernatant was applied to the column which was attached to an amylose resin. After the unbound proteins were washed away, the MBP-BQCV-3CL proteins were recovered by an elution with maltose elution buffer. Concentration of the purified MBPBQCV-3CL proteins was confirmed by Bradford assay with BSA standard (1mg/ml).
RESULTS AND DISCUSSION
Molecular cloning of BQCV-3CL protease fragment
BQCV-3CL protease fragment was cloned into pGEM-3Zf(+)vector and the clone was named as Korean BQCV-3CL protease. Korean BQCV-3CL protease clone was screened by BamHI/SalI restriction analysis (Fig. 2). By the result, this clone may be Black queen cell virus 3CL protease gene. The sequencing result revealed 94.5% homology of nucleotide sequences (Fig. 3) and 99.1% homology of amino acid sequences to a deposited BQCV-3CL protease sequence (Genbank accession No. EF517515.1) (Fig. 4). This result proved that the sequence which was cloned into pGEM-3Zf(+) vector related highly to a Black queen cell virus 3C-like protease sequence published on NCBI.
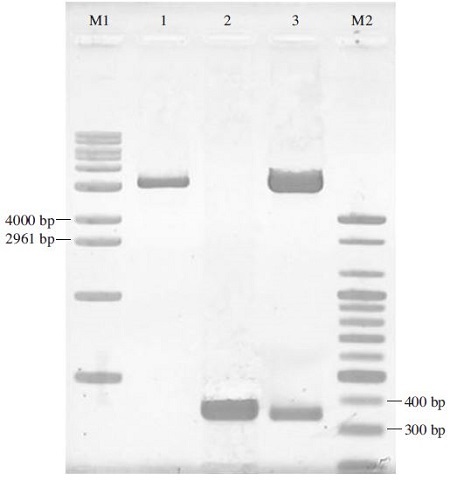
Restriction analysis of pGEM-BQCV-3CL clone by BamHI/SalI. Lane M1 is 1kb DNA ladder (Bioneer, Korea: 500 to10200 bps). Lane M2 is 100bp DNA ladder (Bioneer-Korea: 100 to 2000 bps). Lane 1 is pGEM-3Zf(+) vector which was digested by BamHI/SalI. Lane 2 is PCR product amplified using BQCV-3CL-F-BamHI/BQCV-3CL-R-SalI primer set. Lane 3 is recombinant pGEM-BQCV-3CL protease which was digested by BamHI/SalI. Apparently, the fragments which were digested separately from this clone correspond to an expected PCR product (366 bp) in lane 2 and pGEM-3Zf(+) vector (3179 bp) in lane 1.

Nucleotide sequence homology between Genbank EF517515.1 (Hungary) and Korean BQCV-3CL protease clone. The sequence determined was corresponding 3C-like protease region of EF517515.1 deposited in NCBI databases (nt: 3452...3769) and symbolized by asterisk (*) in the consensus line. Alignment to EF517515.1 revealed 94.5% homology. The sequence in this clone might be Black queen cell virus 3C-like protease sequence.

Amino acid sequence homology between EF517515.1 (Hungary) and Korean BQCV-3CL protease clone. The amino acid sequence of Korean BQCV-3CL protease clone achieved 99.1% homology to Genbank EF517515.1 (a.a: 950...1055) and was marked by asterisk (*) in the consensus line. Obviously, the high homology of amino acid sequence confirmed this clone belongs to Black queen cell virus 3C-like protease.
Estimation of the expression of MBP-BQCV-3CL protein
pMAL-C2 vector, which was constructed as a fusion protein MBP-BQCV-3CL, was transformed into E.coli BL21 (DE3) pLysS. Under a normal condition of induction at 25°C by 0.6mM IPTG during 6hrs, Fig. 5 shows an approximately 63 kDa in protein size and absence in negative controls. The size matched perfectly to an expected protein size, which was calculated from BQCV-3CL protease amino acid sequence. From this result, MBP-BQCV-3CL protein could express well in pMAL-C2 system.
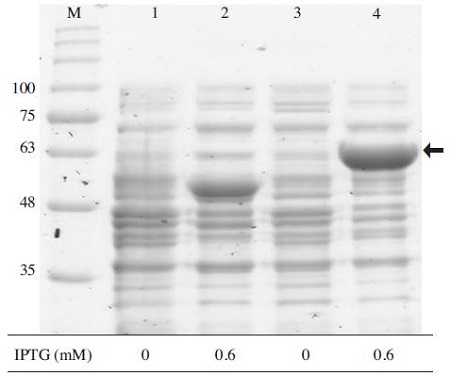
Examination of MBP-BQCV-3CL protein expression in E.coli BL21 (DE3) pLySs. Lane M is protein size marker (GeneDirex, China: 11 to 245 kDa). Lanes 1 and 3 are pMAL-C2 and MBP-BQCV-3CL protein without IPTG, respectively. Lanes 2 and 4 are pMAL-C2 and MBPBQCV-3CL protein induced by IPTG, respectively. After culturing the cell overnight, a subculture step was performed until an optical density reach approximately 0.6 and then the cell were induced by 0.6mM IPTG at 25°C for 6 hours, the expected band around 63kDa was observed clearly on gel, compare to negative controls in lanes 1/2/3. This result confirmed that MBP-BQCV-3CL protein could be expressed well under a normal condition. The black arrow indicated an expected band of molecular weight (63kDa).
Optimal expression conditions of MBP-BQCV-3CL protein
To establish the optimum conditions of subculture, E.coli BL21 (DE3) pLysS containing MBP-BQCV-3CL protein were grown with different optical densities at 600nm (OD600), and then the cells were induced with 0.6 mM IPTG at 25°C for 6 hours. By this induction, 0.6 was determined as an optimized cell density for the expression of MBP-BQCV-3CL protein (Fig. 6). Commonly, 0.6 was the best cell optical density (OD600) for the expresstion in several proteins in E. coli such as Enterovirus A71 3Cprotease (Lin et al., 2012).
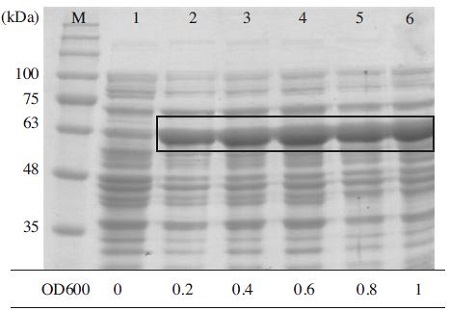
Optimize OD600 value for express MBP-BQCV-3CL protein. MBP-BQCV-3CL proteins in E. coli BL21 (DE3) pLysS were started to induce with 0.6mM IPTG at variable OD600 concentrations. Induced cells were collected after 6 hours. Lane M is protein size marker (GeneDirex, China: 11 to 245 kDa). Lane 1 is a negative control as no induction. Lane 2 to 6 are induced proteins at optical densities 0.2, 0.4, 0.6, 0.8 and 1, respectively. The black box implied the expected protein bands. MBPBQCV-3CL protein expressed highest when the cell density reached to 0.6.
To determine the productivity of recombinant MBPBQCV-3CL protein, different concentration of IPTG was added to the culture when the cell density reached 0.6. The bands that are not intensively shown between IPTG concentration confirmed that 0.1mM IPTG was enough to express MBP-BQCV-3CL protein (Fig. 7). The IPTG concentration for an induction is different between each expression vector systems, the IPTG induction of Bovine viral Diarrhea virus non-structural protein 3 in E. coli occurred vigorously at 0.3mM IPTG (Mahmoodi et al., 2012). In other case, 0.5mM IPTG was identified as an optimization and the protein expression level start dropping at higher IPTG concentration (Nguyen et al., 2012).
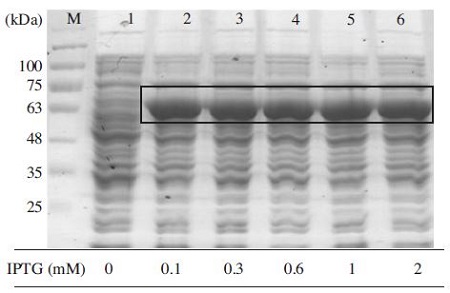
Evaluation of MBP-BQCV-3CL protein depends on IPTG concentration. Lane M is protein size marker (GeneDirex, China: 11 to 245 kDa). Lane 1 is a negative control as no induction. Lane 2 to 6 are inductions at 0.1, 0.3, 0.6, 1 and 2mM IPTG, respectively. By examining variable IPTG concentration, it was clear to confirm that 0.1mM IPTG was enough for express this protein. The black box marked the expected protein bands compare to a negative control in lane 1.
Moreover, the induction time of 6 hours was shown to be optimal (Fig. 8), and it slightly decreased after 12 and 24 hours of incubation. Thus, a long induction time over 6 hours was not necessary to express MBP-BQCV-3CL protein. In addition, lots of protein could express well for short time of IPTG induction (Sankar et al., 1991; No et al., 2011).
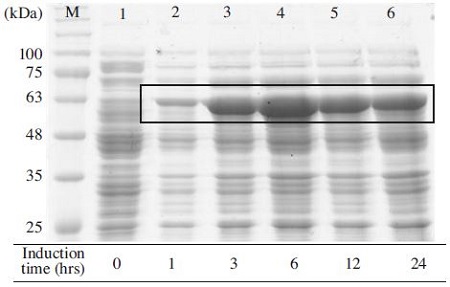
Expression level of MBP-BQCV-3CL protein depended on induction time. The expression of recombinant protein was performed by 10% SDS-PAGE and 1X TGS (Tris/Glycine/ SDS) running buffer. Proteins were heated denaturation before loading on the gel. Induction of MBPBQCV-3CL protein in E.coli BL21 (DE3) pLysS with 0.1mM IPTG was started at 0.6 optical density and cells were collected after different induction time. Lane M is protein size marker marker (GeneDirex, China: 11 to 245kDa). Lane 1 is a negative control without any induction. Lane 2 to 6 are induced proteins at 1, 3, 6, 12 and 24 hours, respectively. The black box implied expected protein bands. This result confirmed that 6 hours was an optimal condition for induction time.
The most crucial factors for protein expression is the induction temperature and whether the expression of MBPBQCV-3CL protein depends on temperature. In this study, 25, 30, and 37°C induction temperatures were examined after fixing all of the conditions. As the result, MBPBQCV-3CL protein expressed at 25°C was better than 30 and 37°C (Fig. 9). Previous researches on protein expression showed that few proteins only could be induced well by IPTG either 37°C (Hata et al., 2000) or 30°C (Zhou et al., 2006). In this case, MBP-BQCV-3CL protein could be easily expressed by different temperatures, it is an advantage for protein manipulation.
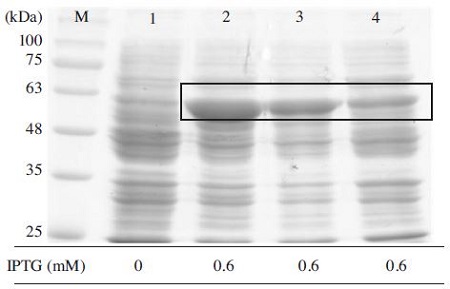
Optimal induction temperature for expression MBPBQCV-3CL protein. Under optimal conditions of OD600, IPTG concentration, time for induction, the evaluation of MBP-BQCV-3CL protein in E. coli BL21 (DE3) pLysS was performed at 25/30/37°C. Lane M is protein size marker marker (GeneDirex, China: 11 to 245 kDa). Lane 1 is a negative control as no induction. Lane 2, 3, 4 are MBP-BQCV-3CL proteins induced at 25/30/37°C, respectively. The black box implied the expected protein bands. In this case, an induction temperature at 25°C was the best for expression.
Purification of MBP-BQCV-3CL protein by amylose resin
As shown in Fig. 10, the MBP-BQCV-3CL protein was recovered at a purified form. After purification, amylose resin will be taken out and saved at 4°C for next purification times. Protein concentration were checked by Bradford assay using BSA (1mg/ml) as a standard. A purified MBP-BQCV-3CL protein yield was calculated approximately 2.22mg per 1ml of cultured cells.
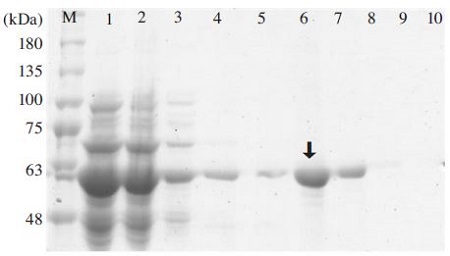
Purification of MBP-BQCV-3CL protein using amylose resin. Lane M is protein size marker (GeneDirex, China: 11 to 245 kDa). Lane 1 is induced total protein. Lane 2 is a part of protein unbound to amylose resin. Lane 3 is a washing by column buffer. Lane 4 is a washing by column buffer in 1% Tween 20. Lane 5 is a washing by column buffer. Lane 6 is an eluted MBP-BQCV-3CL protein. Lane 7 to 10 are column washing. The purified recombinant MBP-BQCV-3CL was shown in lane 6 (Black arrow).
Black queen cell virus is one of pathogens caused the collapse of honeybee population. 3C-like protease in BQCV as well as other Picorna viruses were reported as the most important enzyme which controls the viral replication and capsid assembly by a proteolysis process. A study on Black queen cell virus complete genome sequence found out two specific amino acid sequences existed on BQCV gene where 3C-like protease might be active. In addition, the comparison of peptide sequences which were predicted as specific cleavage sites by 3C-like protease between Black queen cell virus (BQCV) and Plautia stali intestine virus (PSIV), Rhopalosiphum padi virus (RhPV), Droso-phila C virus (DCV) pointed out the conservation of three amino acids surrounding cleavage sites and proved that BQCV was related most closely to PSIV, RhPV, DCV (Leat et al., 2000) (Fig. 1b).
The identification of 3C or 3C-like protease structure attract to many scientists. Greatly, the autocatalytic activity of the 3C protease was examined based on the regulation of the yeast transcriptional activator GAL4 (Dasmahapatra et al., 1992). There were a lot of studies on Foot and Mouth disease virus (FMDV) 3C protease, in which the analysis of FMDV crystallographic structure of 3C protease revealed an overall fold that closely resembles the architecture of trypsin-like serin protease (Zunszain et al., 2009). To date, the majority of research groups world-wide are focusing their drug-discovery efforts on the 3C-like protease enzyme, Japanese sciences succeeded on searching specific inhibitors which inactivated poliovirus 3C protease. Following their result, some serin protease inhibitors, including N-alpha-tosyl-L-lysinyl-chloromethylketone (TLCK) and N-tosyl-L-phenylalaninylchloromethylketone (TPCK), aprotinin and PMSF significantly inhibited the 3CD’ protease activity (Hata et al., 2000). Recently, a new assay for testing inhibitors of picorna virus 3C proteases has been developed. This assay is a cell-based protease which can improve the disadvantages associated with in vitro assays and enables measuring protease activity as well as the inhibitory activity of compounds in a cellular environment (Linden et al., 2014).
In our study, we cloned BQCV-3CL protease gene in pGEM-3Zf(+) vector. BQCV-3CL protease clone revealed 94.5% homology of nucleotide sequence (Fig. 3) and 99.1% homology of amino acid sequence to a deposited sequence (Genbank accession No. EF517515.1) (Fig. 4). The accession number, KP307016, was conferred by NCBI for BQCV-3CL protease gene confirmation in this study. BQCV-3CL protease gene was constructed in pMAL-C2 vector, and it was successfully expressed in E. coli BL21 (DE3) pLysS under an optimal induction at 0.6 cell optical density with 0.1mM IPTG for 6 hours at 25°C. This condition was the same as the previous research (Lee et al., 2014). On the other hand, another research on 3C protease suggested that the induction at 37°C for 3 hours is an optimal condition (Hata et al., 2000). Finally, MBPBQCV-3CL protein, which contained MBP-tag at Nterminal, was recovered at high efficiency (2.22mg per 1ml of cultured cells). Therefore, we could prepare a potential antigen source for generating monoclonal antibodies and develop a rapid Kit for sensitive detection of BQCV in honeybees. Nevertheless, the characteristics and functions of BQCV-3CL protease is an attractive direction for future studies.
Acknowledgments
This work was supported by Bio-Industry Technology Development Program (312027-3) and Advanced Production Technology Development Program (112042-03) .
LITERATURE CITED
-
Allaire, M., M. M. Chernaia, B. A. Malcolm, M. N. James, (1994), Picornal viral 3C cysteine protease have a fold similar to chymotrypsin-like serin protease, Nature, 369, p72-76.
[https://doi.org/10.1038/369072a0]

-
Bailey, L., R. D. Woods, (1977), Two more small RNA viruses from honey bees and further observations on Sacbrood and Acute bee-paralysis viruses, J. Gen. Virol, 37, p175-182.
[https://doi.org/10.1099/0022-1317-37-1-175]

-
Cui, S., J. Wang, T. Fan, B. Qin, L. Guo, X. Lei, M. Wang, Q. Jin, (2011), Crystal structural of human enterovirus 71 3C protease, J. Mol. Biol, 408, p449-461.
[https://doi.org/10.1016/j.jmb.2011.03.007]

-
Curry, S., N. R. Rosell, P. A. Zunszain, R. J. Leatherbarrow, (2007), Foot and mouth disease virus 3c protease: Recent structural and functional insights into an antiviral target, J. Biol. Chem, 39, p1-6.
[https://doi.org/10.1016/j.biocel.2006.07.006]

-
Dasmahapatra, B., B. Didomenico, S. Dwyer, J. Ma, I. Sadowski, and J. Schwartz, (1992), A genetic system for studying the activity of a proteolytic enzyme, Pro. Natl. Acad. Sci. USA, 89, p4159-4162.
[https://doi.org/10.1073/pnas.89.9.4159]

- Dougherty, W. G., and B. L. Semler, (1993), Expression of virusencoded proteinase: functional and structural similarities with cellular enzymes, Microbiol. Rev, 57, p781-822.
-
Gorbalenya, A. E ., A. P. Donchenko, V. M. Blinov, (1986), Poliovirus-encoded proteinase 3C: a possible evolutionary like between serine and cysteine protease families, FEBS Lett, 194, p253-257.
[https://doi.org/10.1016/0014-5793(86)80095-3]

-
Hata, S ., T. Sato, H. Sorimachi, S. Ishiura, K. Suzuli, (2000), A simple purification and fluorescent assay method of the poliovirus 3C protease searching for specific inhibitors, 84, p117-126.
[https://doi.org/10.1016/S0166-0934(99)00138-X]

-
Le Gall, O., P. Christian, C. M. Fauquet, A. M. King, N. J. Knowles, G. Nakashima, N. Stanway, and A. E. Gorbalenya, (2008), Picornavirales, a proposed order of positive-sense single-strandedRNA viruses with a pseudo-T = 3 virion architecture, Arch. Virol, 153, p715-727.
[https://doi.org/10.1007/s00705-008-0041-x]

- Leat, N., B. Ball, B. Govan, and S. Davison, (2000), Analysis of the complete genome sequence of black queen cell virus, a picorna-like virus of honey bees, Journal of General Virology, 81, p2111-2119.
- Lee, J. S., H. Y. Lim, and B. S. Yoon, (2014), Over-expression and purification of Peptidase-C3G protein of Deformed wing virus, Korean J. Apiculture, 29(1), p57-62.
-
Lin, Y. J., Y. C. Chang, N. W. Hsiao, J. L. Hsieh, C. Y. Wang, S. H. Kung, F. J. Tsai, Y. C. Lan, and C. W. Lin, (2012), Fisetin and rutin as 3C protease inhibitors of enterovirus A71, J. Virol. Methods, 182, p93-98.
[https://doi.org/10.1016/j.jviromet.2012.03.020]

-
Linden, V. D. L., R. Ulferts, S. B. Nabuurs, Y. Kusov, H. Liu, S. George, C. Lacroix, N. Goris, D. Lefebvre, K. H. W. Lanke, K. D. Clercq, R. Hilgenfeld, J. Neyts, and F. J. M. V. Kuppeveld, (2014), Application of a cell-based protease assay for testing inhibitors of picornavirus 3C proteases, J. Antiviral research, 103, p17-24.
[https://doi.org/10.1016/j.antiviral.2013.12.012]

- Mahmoodi, P., M. R. S. A. Shapouri, M. Ghorbanpour, M. R. H. Hajikolaei, M. Lotfi, and M. Ekhtelat, (2012), Jundishapur J, Micro, 6(7), pe7044.
-
Malcolm, B. A., (1995), The picornaviral 3C proteases: cysteine nucleophiles in serine protease folds, Protein Sci, 4, p1439-1445.
[https://doi.org/10.1002/pro.5560040801]

- MayO, M. A., (2002), Virus taxonomy-Houston 2002, Arch Virol, 147, p1071-1076.
- Nguyen, V. P., B. Lee, and B. S. Yoon, (2012), Over-expression and purification of capsid protein of Sacbrood virus in E. coli, Korean J. Apiculture, 27(3), p209-214.
- No, J. N., S. H. Han, and B.S. Yoon, (2011), Capsid protein gene expression of Israel Acute Paralysis virus in Honeybee (Apis mellifera L.), Korean J. Apiculture, 26(3), p221-227.
-
Palmenberg, A.C., (1990), Proteolytic processing of picornaviral polyprotein, Annu. Rev. Microbiol, 44, p603-623.
[https://doi.org/10.1146/annurev.mi.44.100190.003131]

- Sankar, S, and A. G. Porter, (1991), Expression, purification, and properties of recombinant Encephalomyocarditis virus RNA-Dependent RNA polymerase, J. Virol, 65, p2993-3000.
-
Shan, Y., H. Xia, C. Dong, Z. Cheng, X. Xia, J. Zhang, X. Zhou, and Y. Hu, (2012), Identification and characterization of Iflavirus 3C-like protease processing activities, J. Virol, 428, p136-145.
[https://doi.org/10.1016/j.virol.2012.04.002]

-
Zhou, L., J. Zhang, X. Wang, H. Jiang, F. Yi, and Y. Hu, (2006), Expression and characterization of RNA-dependent RNA polymerase of Dendrolimus punctatus Tetravirus, J. BioChem, 39, p571-577.
[https://doi.org/10.5483/bmbrep.2006.39.5.571]

-
Zunszain, P. A., S. R. Knox, T. R. Sweeney, J. Yang, N. R. Rosell, G. J. Belsham, R. J. Leatherbarrow, and S. Curry, (2009), Insights into cleavage specificity from the crystal structure of Foot-and-Mouth disease virus 3C protease complexed with a peptide substrate, J. Mol. Biol, 395, p375-389.
[https://doi.org/10.1016/j.jmb.2009.10.048]

