
Development of On-site Molecular Diagnostics of Nosema diseases in Honeybee
Abstract
Nosema is a parasitic microsporidia fungal disease. There are now two different types of Nosema: N. apis and N. ceranae. Nosema affects adult bees only by infecting epithelial cells lining the mid-gut after spores are ingested. In this study, the LAMP (Loop-mediated Isothermal amplification) detection method againsts Nosema diseases which can be easily recognized its result by naked eye instead of electrophoresis was developed. Nucleic acid fluorescent dye, SYBR Green-and Gene-FinderTM‚ were adopted in this method and real-time thermal cycler and UF-PCR equipment were also used. The new method can perform the amplification of target DNA successfully within 30 minutes and againsts each pathogen by a successfully amplification from 1×102 molecules of template DNA with quantitative manner in real-time. The new method was demonstrated to be a specific, sensitive and easy tool for on-site detection. Thus, it may be useful for the monitoring of natural infection in N. apis and N. ceranae.
Keywords:
Nosema apis, Nosema ceranae, LAMP, DetectionINTRODUCTION
Nosemosis is caused by the obligate intracellular and spore-forming parasites, Nosema apis and Nosema ceranae, which attack the lining of the middle intestine of the worker, queen, and drones of honeybee (Bailey, 1955). N. apis infecting Apis mellifera was first described in 1907 by Enock Zander and to be considered as the perpetrator of Nosema disease in honeybee A. mellifera for a long time (Higes et al., 2007). N. apis is generally considered to be not severe pathogenic, but be prevalent throughout the world (Fries et al., 1984; Anderson and Giacon, 1992). On the other hand, N. ceranae infecting Apis cerana was first discovered in 1994 (Fries et al., 1996).
Nosema is transmitted from the worker bees feed on exchanges (Smith, 2012). Nosemosis can induce queen supercedure (Webster et al., 2004), reduce pollen collection (Anderon and Giacon, 1992), and shorten bee life span (Wang and Moeller, 1970).
According to the detection result about the twelve kinds of honeybee disease published by Korea animal and plant quarantine agency in 2013, out of which the occurrence of Nosema disease was found the highest level (36.49%). The timing of Nosema diagnosis is very important, because Nosema spp. infestation is associated with reduced honey production and increased mortality in winter (Higes et al., 2006, 2007) and lead to shorten the worker life span and considerable weakening of heavily infected colonies, resulting in significant economic damage (Fries et al., 1984; Anderson and Giacon, 1992).
Nosema disease diagnostic methods using PCR has already been developed (Lim et al., 2014; Byeon et al., 2014; Yoo et al., 2011). However, the detection of disease in laboratory takes a long time and on-site detection is difficult because these detection methods need the expensive equipment. Also no method is available to distinguish between N. apis and N. ceranae.
Loop-mediated Isothermal Amplification (LAMP) method developed in 2000 has a high specificity and sensitivity as a new method for quickly amplifying a nucleic acid in isothermal conditions (Notomi et al., 2000). LAMP method has three main features. Firstly, all reactions are carried out under isothermal conditions. Compared to PCR and real-time PCR assays, LAMP is requiring only heat block or regular laboratory water bath for reaction instead of expensive equipment like thermocycler (Ushikubo, 2004). Secondly, the amplification efficiency is extremely high, resulting in a tremendous amount of amplification products (Yamazaki et al., 2008). Thirdly, the method has a high specificity (Yamazaki et al., 2009).
GenecheckerTM device (Dongwoo Science Company, Korea) developed in 2013 incorporates the high level of real-time fluorescence detection technology and it is possible to examine the reaction results through a window of the above equipment. The equipment is also small and easy to carry. By using 12V DC voltage, this equipment is able to be connected to the vehicle battery or power source and on-site analysis is possible.
In this study, by combining the advantages of the LAMP method and the GenecheckerTM, the new on-site molecular diagnostic method was developed that is capable to detect N. apis and/or N. ceranae separately.
MATERIALS AND METHODS
Infected honeybee
All infected honeybees and larvae were collected from apiaries in whole of 2014 from Jeju and Ulsan, Korea. These samples were stored at -70°C until processed.
Genomic DNA extraction
Larvae and adult honeybee were homogenized at 6000 rpm for 60 sec by MagNa Lyser (Roche, Switzerland) and genomic DNA of each sample was extracted using AccuPrep® Genomic DNA Extraction Kit (Bioneer Inc., Korea). The experiments were carried out according to the instructions of the manufacturer. The final concentration of total genomic DNA was determined using a spectrophotometer (Effendorf, Germany) before applying to the LAMP reaction. All genomic DNA was stored at -70°C until processed.
Template DNAs preparation
pBX-Nosema apis (Lim et al., 2014) and pBX-Nosema (Yoo et al., 2008) were used as standard templates. pBX-Nosema apis includes the sequence that shows 99% homology with N. apis sequence (GenBank No.U97150). pBX-Nosema includes the sequence that show 99% homology with Nosema ceranae sequence (GenBank No.DQ486027). Each clone used to template for optimized condition and test the sensitivity of specific LAMPs.
Specific primer sets for LAMP
N. apis-specific LAMP primers were designed based on the sequence of large subunit ribosomal RNA of N. apis (GenBank No.U97150, 1860-2320nt). Specific primer sets were consisting of four primers: two outer primers (N. apis F3 and N. apis B3) and two inner primers (N. apis FIP consisting of F2 and F1c sequences and N.apis BIP consisting of B2 and B1c sequences) (Fig. 1 and Table 1). All primers were synthesized by Bionics Corporation, Korea.
N.ceranae-specific LAMP primers were designed by Lee et al. (2010). They were Nosema F3 (25nt), Nosema B3 (19nt), Nosema FIP (45nt), and Nosema BIP (48nt) (Table 1)
Optimization of temperature for LAMP reaction
Reaction were performed in a total 25μl volume in case of N. apis-specific LAMP, containing 20pmole each of N-apis F3, N-apis B3, 80pmole each of N-apis FIP, N-apis BIP, 10mM dNTPs, 8 units of Bst DNA polymerase large fragment (New England Biolabs), 10X ThermoPol Reaction Buffer (20mM Tris-HCl, 10mM KCl, 10mM (NH4)2SO4, 2mM MgSO4, 0.1% Triton X-100), and template DNA. The mixture was incubated for 60 min at 41.5~57.0°C and the reaction was terminated by heating to 85°C for 10 min. After LAMP reaction, the amplicons were confirmed by agarose-gel electrophoresis.
Optimization of condition for LAMP reaction
In order to establish the optimal reaction conditions for N. apis-LAMP, the optimum conditions of the LAMP components were confirmed. The ratio of the outer primer and inner primer (1:4) showed the highest efficiency (Notomi et al., 2000). This ratio is maintained and the reactions were examined by changing only the absolute concentration of the primer. In other words, when the concentration of each outer primer was 5pmole, 10pmole, 15pmole, 20pmole, 25pmole, the concentration of each inner primers was 20pmole, 40pmole, 60pmole, 80pmole, 100pmole. To optimize the concentration of dNTP, the reactions were examined by using the each dNTP that was set as 2.5mM, 5.0mM, 7.5mM, 10.0mM, 12.5mM, 15.0mM, and to optimize concentration of BST polymerase, each 4U, 8U, 12U, 16U, 20U of the Bst polymerase was used in LAMP reaction.
Construction of the Loop primers
The addition of Loop primers might accelerate the LAMP reaction (Nagamine et al., 2002). Each loop primer set for N.apis-specific LAMP and N.ceranae-specific LAMP was designed. The loop primer set for N.apis-specific LAMP was designed based on N. apis large subunit ribosomal RNA gene (GenBank No.U97150). Forward loop primer called N. apis Loop F was designed to locate between the F2C and F1C that were complementary sequence of N. apis sense sequence. And reverse loop primer called N. apis Loop B was designed to locate between the B1C and B2C that were complementary sequence of N. apis anti-sense sequence (Fig. 4A).
The loop primer set for N. ceranae-specific LAMP was designed based on Nosema ceranae small subunit ribosomal RNA gene (GenBank No.DQ486027). Forward Loop primer called Nosema Loop F was designed to locate between the F2C and F1C that were complementary sequence of N. ceranae sense sequence. And reverse loop primer called Nosema Loop B was designed to locate between the B1C and B2C that were complementary sequence of N. ceranae anti-sense sequence (Fig. 4B).
Determination of reaction time through the Realtime LAMP assay
To determine the N. apis-specific LAMP reaction time according to the DNA molecules for N. apis and the N. ceranae-specific LAMP reaction time according to the DNA molecules for N. ceranae, each of the real-time LAMP was carried out using 10-fold serial dilution of each pBX-Nosema apis (Lim et al., 2014) and pBX-Nosema (Yoo et al., 2008). One cycle is set to one minute, and reactions were incubated at 54.0°C for 70 min and 85.0°C for 10 min. After the LAMP reaction, the results were confirmed by electrophoresis analysis.
LAMP reaction with GenecheckerTM
The LAMP reactions were carried out under the modified standard conditions for 10ul. All LAMP reactions were conducted on a Model UF-100 GenecheckerTM system with polymer chip named Rapi:chip (Dongwoo Science, Seoul, Korea).
Sensitivity of LAMP reaction with GenecheckerTM
To evaluate the detection limit of N. apis-specific LAMP with GenecheckerTM, LAMP reaction was performed under the standard conditions. pBX-Nosema apis (Lim et al., 2014) was serially diluted 10-fold from 1×108 molecules and 1μl of each dilution was used as a template for the N. apis-specific LAMP reaction.
And also, to determine the detection limit of N. ceranae-specific LAMP with GenecheckerTM, LAMP reaction was performed under the standard conditions. pBX-Nosema (Yoo et al., 2008) was serially diluted 10-fold from 1×108 molecules and 1μl of each dilution was used as a template for the N. ceranae-specific LAMP reaction.
Specificity of LAMP reaction with GenecheckerTM
To assess the specificity of the N. apis-specific primer sets (N. apis F3, N.apis B3, N. apis FIP, N. apis BIP, N. apis Loop F, N. apis Loop B) and N. ceranae-specific primer sets (Nosema F3, Nosema B3, Nosema FIP, Nosema BIP, Nosema Loop F, Nosema Loop B), LAMP reactions were performed under a standard condition using genomic DNA from honeybee sample infected by A. apis, A. flavus and N. ceranae.
Fluorescent dye test
LAMP products were visualized under daylight by yellow fluorescence after the addition of Gene-FinderTM (Baygene Biotech Company Limited, China), under UV by green fluorescence after the addition of SYBR GreenⅠ.
RESULTS AND DISCUSSION
Designing of specific primer sets for LAMP
The nucleotide sequence of the sense strand of the N. apis genome is shown. DNA sequences used for primer design are underlined (Fig. 1).
Optimal temperature of LAMP reaction
To determine the optimal temperature for the N. apis-specific LAMP assay, pBX plasmid containing a part of the N. apis large subunit ribosomal RNA gene was used. The each LAMP reaction was performed under isothermal conditions within 41.5, 44.7, 47.2, 50.0, 54.3, and 57.0°C for 60 min. After LAMP reaction, the amplicons were confirmed by agarose-gel electrophoresis. The amplicons were formed between 41.5°C and 54.3°C. For simultaneous diagnosis with N. ceranae, 54°C was chosen as the optimum reaction temperature for the subsequent LAMP assays (Fig. 2).
The optimum temperature for N.ceranae-LAMP was set as 54.0°C.
Optimal conditions of LAMP reaction
In order to establish the optimal reaction conditions for N. apis-specific LAMP, several concentrations of each composition (Primers, dNTP, Bst polymerase) were tested. In case of primers, the strongest amplicon was formed when the inner primer was 80pmole and outer primer was 20pmole. In spite of repeated experiments, the optimum concentration of the inner primer was determined to 80pmole and the optimum concentration of the outer primer was determined to 20pmole (Fig. 3A)
In order to determine the optimal concentration of dNTP for N. apis-specific LAMP, each dNTP (2.5mM, 5.0mM, 7.5mM, 10.0mM, 12.5mM and 15.0mM ) was added to each reaction solution.
After specific LAMP, agarose-gel electrophoresis was performed. The strongest amplicon was formed when the dNTP concentration was 10mM. Therefore, the optimum concentration of dNTP was determined to 10mM (Fig. 3B).
Moreover, the determination of optimal concentration of Bst polymerase for N. apis-specific LAMP was performed. Various units of Bst polymerase (4U, 8U, 12U, 16U and 20U) were added to each reaction. After the reaction, the amplicons were confirmed by agarose-gel electrophoresis. N. apis-specific LAMP products could be amplified between 4U to 16U. The various sizes of the products were observed when 8U Bst polymerase was added to LAMP reaction solution. Therefore, the optimal concentration of Bst DNA polymerase was determined to 8U (Fig. 3C).
The N. apis-specific LAMP standard condition was set in a total volume of 25ul containing 20pmole each of outer primers, 80pmole each of inner primers, 10mM dNTP, 8U Bst polymerase large fragment (New England Biolabs), 10X TermolPol Reaction buffer (20mM Tris-HCl, 10mM KCl, 10mM (NH4)2SO4, 2mM MgSO4, 0.1% Triton X-100), and template DNA. Then, the mixture was incubated at 54.0°C for 60 min.
The standard condition for N. ceranae-LAMP was set in a total volume of 25ul containing 5pmole each of outer primers, 20pmole each of inner primers, 5mM dNTP, 4U Bst polymerase large fragment (New England Biolabs), 10X TermolPol Reaction buffer (20mM Tris-HCl, 10mM KCl, 10mM (NH4)2SO4, 2mM MgSO4, 0.1% Triton X-100), and template DNA. Then, the mixture was incubated at 54.0°C for 60 min.
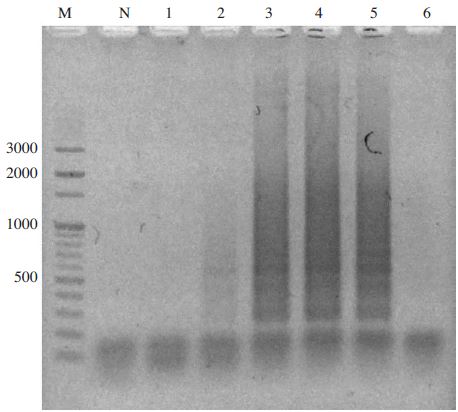
Temperature gradient N. apis-specific LAMP. Lane M1 is 100 bp ladder Marker (Bioneer). Lane 1 to 6 were specific LAMP products from each LAMP under isothermal temperature at 41.5, 44.7, 47.2, 50.0, 54.3, and 57.0°C, respectively. Lane N is negative control without template. Optimal elongation temperature was determined at 54.0°C.
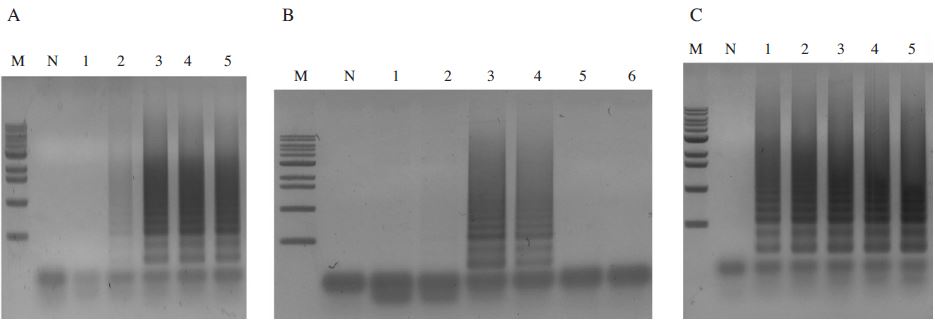
The optimal concentration of reaction solutions in N. apis-specific LAMP. Panel A. The optimal concentration of primers. Lane M is 1kb ladder marker (Bioneer); 10.2, 8, 5.9, 5, 4, 2.9, 2, 1.6, 1, 0.5kb, respectively. Lane N, 0pmole N.apis-FIP/BIP and 0pmole N.apis-F3/B3; Lane 1, N. apis-specific LAMP using 20pmole N.apis-FIP/BIP and 5pmole N. apis-F3/B3; Lane 2, N. apis-specific LAMP using 40pmole N. apis-FIP/BIP and 10pmole N. apis-F3/B3; Lane 3, N. apis-specific LAMP using 60pmole N. apis-F3/B3 and 15pmole N. apis-F3/B3; Lane 4, N. apis-specific LAMP using 80pmole N.apis-FIP/BIP and 20pmole N. apis-F3/B3; Lane 5, N. apis-specific LAMP using 100pmole N. apis-FIP/BIP and 25pmole N. apis-F3/B3. The optimal concentration of primer was determined at 80pmole N. apis-FIP/BIP and 20pmole N. apis-F3/B3. Panel B. The optimal concentrations of dNTP. Lane M is 1kb ladder marker (Bioneer); 10.2, 8, 5.9, 5, 4, 2.9, 2, 1.6, 1, 0.5kb, respectively. Lane N is negative control. Lane 1 to 6, LAMP products using 2.5mM; 5mM; 7.5mM; 10mM; 12.5mM; 15mM dNTP. The optimal concentration of dNTP was determined at 10mM. Panel C. The optimal concentration of Bst polymerase. Lane M is 1kb ladder marker (Bioneer); 10.2, 8, 5.9, 5, 4, 2.9, 2, 1.6, 1, 0.5kb, respectively. Lane N is negative control. Lane 1 to 5, LAMP products using 4U; 8U; 12U; 16U; 20U Bst polymerase. The optimal concentration of Bst polymerase was determined at 8U.
Specific Loop primers sets for LAMP
According to the result that the addition of Loop primers can accelerate the LAMP reaction of the paper (Nagamine et al., 2002), each loop primer set for N. apis-specific LAMP and N. ceranae-specific LAMP was designed (Fig. 4 and Table 2).
During the same time, the LAMP reactions were performed and the results were confirmed by agarose-gel electrophoresis. As a result, the lane added Loop primer was working well than the lane not added loop primer (data not shown).
Reaction time of LAMP with real-time LAMP assay
In order to determine the reaction time of N. apis-specific LAMP according to the number of DNA molecules, pBX-Nosema apis (Lim et al., 2014) was serially diluted 10-fold, and 1μl of each dilution was used as a template for the real-time LAMP reaction. As a result, at least 1×104 molecules of DNA could be detected within 35 min (Fig. 5A). In addition, it was confirmed again by electrophoresis analysis (data not shown).
To determine the N. ceranae-specific LAMP reaction time according to the DNA molecules for N. ceranae, the real-time LAMP was carried out using 10-fold serial dilution of pBX-Nosema (Yoo et al., 2008). After the LAMP reaction, at least 1×105 molecules of DNA could be detected within 30 min (Fig. 5B). The results were confirmed again by electrophoresis analysis (data not shown).
Detection of LAMP product with GenecheckerTM
After amplification, the well containing template DNA showed clearly green fluorescence signal. However, the well without template didn't show fluorescence signal (Fig. 6). The results were confirmed again by electrophoresis analysis.
The sensitivity of LAMP assay with GenecheckerTM
In case of N. apis-specific LAMP, the fluorescence signals were gained in the range of 108 to 102 molecules of pBX-Nosema apis (Fig. 7A). However, in case of N. ceranae-specific LAMP, the fluorescence signals were gained in the range of 108 to 103 molecules of pBX-Nosema (Fig. 7B). The results were confirmed again by electrophoresis analysis.
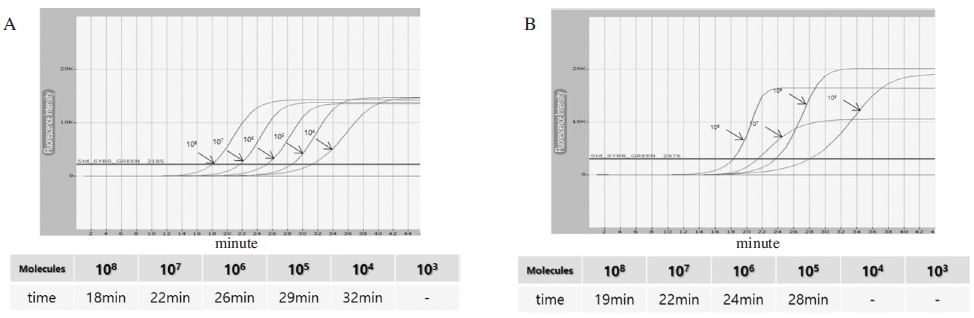
The LAMP result for detection time in accordance with the number of DNA molecules using Real-time fluorescent measurement. Panel A. The fluorescence results of N. apis-specific LAMP using the 10-fold serial dilutions from 1×108 molecules. It is possible to diagnose the N. apis 1×104 molecules within 35 minutes. The R2 value of standard curve was 0.984 (data not shown) Panel B. The fluorescence results of N. ceranae-specific LAMP using the 10-fold serial dilutions from 1×108 molecules. It is possible to diagnose the Nosema ceranae 1×105 molecules within 30 minutes. The R2 value of standard curve was 0.984 (data not shown).
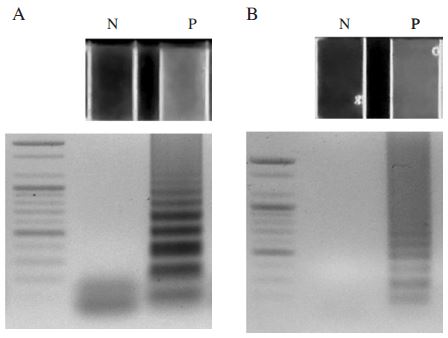
The results of the Nosema-specific LAMP. Panel A. The N. apis-specific LAMP results using GeneckeckerTM and agarose gel electrophoresis. Lane N is Negative control without template. Lane P is positive control using pBX-Nosema apis plasmid DNA (1ng). Panel B. The N. ceranae-specific LAMP results using GeneckeckerTM and agarose gel electrophoresis. Lane N is Negative control without template. Lane P is positive control using pBX-Nosema plasmid DNA (1ng).
Species specificity of LAMP with GenecheckerTM
In this study, genomic DNAs from honeybees infected by A. apis, A. flavus and N. ceranae were tested to evaluate the specificity of the N. apis-specific LAMP primer sets (N. apis F3, N. apis B3, N. apis FIP, N. apis BIP, N. apis Loop F, N. apis Loop B) and the N. ceranae-specific LAMP primer sets (Nosema F3, Nosema B3, Nosema FIP, Nosema BIP, Nosema Loop F, Nosema Loop B). The N. apis-specific LAMP primers demonstrated a high degree of specificity for N. apis by amplifying N. apis but yielding negative results with all of the other pathogens tested (Fig. 8A). On the other hand, the N. ceranae-specific LAMP primers demonstrated a high degree of specificity for N. ceranae by amplifying N. ceranae but yielding negative results with all of the other pathogens tested (Fig. 8B).
Detection of LAMP product with fluorescent dye
When Gene-FinderTM was added to the tubes after the reaction, yellow fluorescence was observed with naked dye in a positive reaction, whereas orange fluorescence was observed in a negative reaction (Fig. 9A). Meanwhile, when SYBR Green I was added to the tubes after the reaction, green fluorescence was observed with the naked eye in a positive reaction, whereas no signal was observed in a negative reaction (Fig. 9B).
Nosema spp. are a microsporidian, a small, unicellular parasite recently reclassified from protozoa to the Fungi cluster, rank Opisthokonta (Adl et al., 2005) that are ubiquitous exhibiting an extensive range of hosts including honey bees. In many papers previously published, N. ceranae has jumped host from A. cerana to A. mellifera and become distributed almost worldwide (Klee et al., 2007). A quick and accurate molecular genetic method of detection of microsporidia is important for identification (Klee et al., 2006) because species of microsporidia are often difficult to distinguish using morphological criteria (Larsson, 1986; Weiss and Vossbrinck, 1999).
In this study, new LAMP methods for the specific detection of N. apis and N. ceranae in honeybees were developed and are termed as a N. apis-specific LAMP and N. ceranae-specific LAMP. In case of N. apis-specific LAMP, six primers for N. apis specific LAMP (N. apis F3, N. apis B3, N. apis FIP, N. apis BIP, N. apis Loop F and N. apis Loop B) were designed based on N. apis large subunit ribosomal RNA gene (GenBank, Accession no.U97150.1). The conditions of the LAMP reaction were optimized by an incubation of the target DNAs; pBX-Nosema apis with the six primers at 54°C for 60 min. Moreover, the diagnostic method using GENECHECKERTM‚ system allowed a direct observation of results by naked eye instead of electrophoresis. The new method has an advantage that can successfully perform amplification of 104 molecules target DNA within 30 minutes. And specific N. apis primer set is able to achieve specific detection of N. apis whereas no amplification to other pathogen like Ascosphaera apis, Aspergillus flavus and Nosema ceranae.
In case of N. ceranae-specific LAMP, the N. ceranae-specific LAMP method (Lee et al., 2010) was modified. Six primers for N. ceranae-specific LAMP (Nosema F3, Nosema B3, Nosema FIP, Nosema BIP, Nosema Loop F and Nosema Loop B) were designed based on Nosema ceranae small subunit ribosomal RNA gene (GenBank, Accession no.DQ486027). The conditions of the LAMP reaction were optimized by an incubation of the target DNAs; pBX-Nosema with the six primers at 54°C for 60 min. Moreover, the diagnostic method using GenecheckerTM system allowed a direct observation of results by naked eye instead of electrophoresis. This method against Nosema ceranae can be successfully amplified from 1× 103 molecules of template DNA. Specific Nosema ceranae primer set is able to achieve specific detection of Nosema ceranae whereas no amplified to other pathogen like A. apis, A. flavus and N. apis.
By using GenecheckerTM that adopts DC 12V power input and can be connected to a general car power source, these LAMP methods may be expected to be useful tool for rapid, specific detection of Nosema diseases in the field and for monitoring of natural infection in A. mellifera and A. cerana.
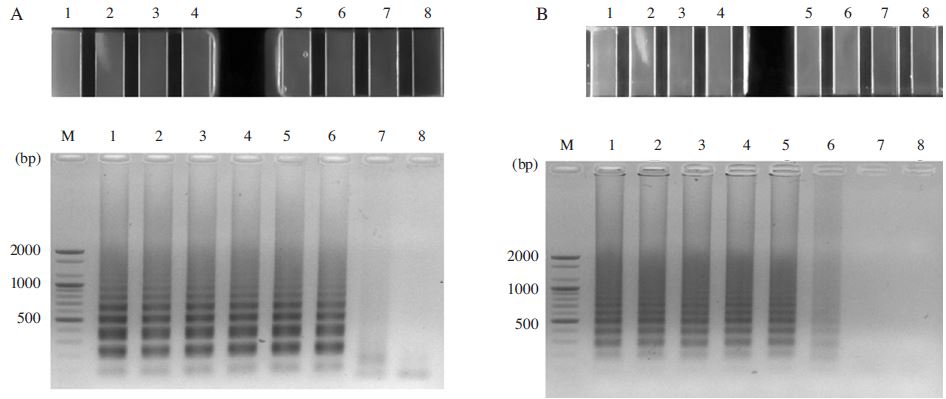
Sensitivity test of the Nosema-specific LAMP. Panel A. The N. apis-specific LAMP results using GenecheckerTM (up) and agarose gel electrophoresis (down). Lane M is 100bp DNA ladder (Bioneer). Lane 1, N. apis-specific LAMP using 1×108 molecules. Lane 2, N. apis-specific LAMP using 1×107 molecules. Lane 3, N. apis-specific LAMP using 1×106 molecules. Lane 4, N. apis-specific LAMP using 1×105 molecules. Lane 5, N. apis-specific LAMP using 1×104 molecules. Lane 6, N. apis-specific LAMP using 1×103 molecules. Lane 7, N. apis-specific LAMP using 1×102 molecules. Lane 8 is negative control without template. Panel B. The N. ceranae-specific LAMP results using GenecheckerTM (up) and agarose gel electrophoresis (down). Lane M is 100bp DNA ladder (Bioneer). Lane 1, N. ceranae-specific LAMP using 1×108 molecules. Lane 2, N. ceranae-specific LAMP using 1×107 molecules. Lane 3, N. ceranaee-specific LAMP using 1×106 molecules. Lane 4, N. ceranae-specific LAMP using 1×105 molecules. Lane 5, N. ceranae-specific LAMP using 1×104 molecules. Lane 6, N. ceranae-specific LAMP using 1×103 molecules. Lane 7, N. ceranae-specific LAMP using 1×102 molecules. Lane 8 is negative control without template.
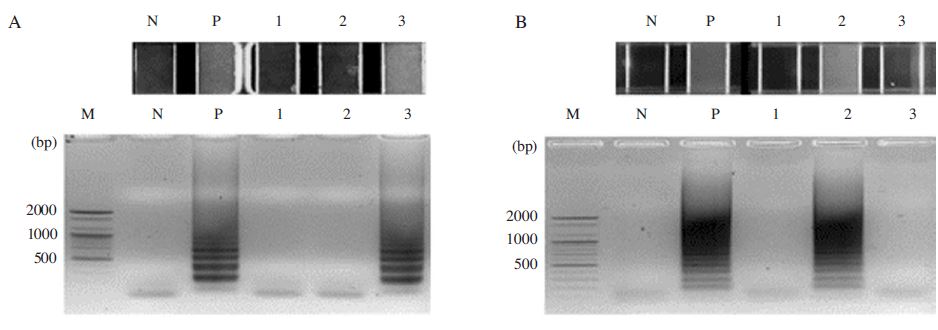
Detection of N. apis-specific LAMP and N. ceranae-specific LAMP. As target templates genomic DNA isolated from honeybee sample. Each sample diagnosed A. apis, A. flavus, and N. ceranae infection were used for the experiment. Panel A. LAMP reaction with N. apis LAMP primer sets to Honeybee genomic DNA. Lane M is DNA size marker; 2, 1.6, 1.2, 1, 0.9, 0.8, 0.7, 0.6, 0.5, 0.4, 0.3, 0.2, 0.1kb, respectively. Lane N is no template as a negative control, Lane P is reacted with pBX-Nosema apis, Lane1 is reacted with Honeybee genomic DNA that was infected by A. apis and A. flavus. Lane 2 is reacted with Honeybee genomic DNA that was infected by N. ceranae. Lane 3 is reacted with artificial Honeybee genomic DNA that was infected by N. apis. Panel B. LAMP reaction with Noseme ceranae LAMP primer sets to Honeybee genomic DNA. Lane M is DNA size marker; 2, 1.6, 1.2, 1, 0.9, 0.8, 0.7, 0.6, 0.5, 0.4, 0.3, 0.2, 0.1kb, respectively. Lane N is no template as a negative control, Lane P is reacted with pBX-Nosema, Lane1 is reacted with Honeybee genomic DNA that was infected by A. apis and A. flavus. Lane 2 is reacted with Honeybee genomic DNA that was infected by N. ceranae. Lane 3 is reacted with artificial Honeybee genomic DNA that was infected by N. apis.
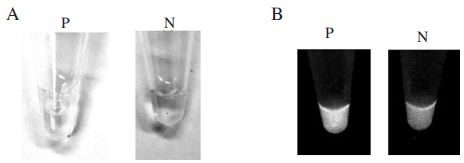
Gene-FinderTM Nucleic acid fluorescent dye and SybrGreenⅠ test of N. apis-specific LAMP. Panel A. LAMP product (P) and negative control (N) were stained by Gene-FinderTM Nucleic acid fluorescent dye. Panel B. LAMP product (P) and negative control (N) were stained by SybrGreenⅠ. Each product was observed by ultraviolet transilluminator.
Acknowledgments
This work was supported by Kyonggi University Research Grant 2014.
References
-
Adl, S. M., A. G. B. Simpson, M. A. Farmer, R. A. Andersen, O. R. Anderson, J. R. Barta, et al , (2005), The new higher level classification of Eukaryotes with emphasis on the taxnomy of Protists, J. Eukaryot Microbiol, 52, p399-451.
[https://doi.org/10.1111/j.1550-7408.2005.00053.x]

-
Anderson, D. L., H. Giacon, (1992), Reduced pollen collection by honeybee (Hymenoptera, Apidae) colonies infection with Nosema apis and sacbrood virus, J. Econ. Entomol, 85, p47-51.
[https://doi.org/10.1093/jee/85.1.47]

-
Bailey, L., (1955), The infection of the ventriculus of the adult honeybee by Nosema apis (Zander), Parasitology, 45, p86-94.
[https://doi.org/10.1017/S0031182000027451]

-
Byeon, G. H., Tran Van Toan, M. Y. Lee, H. S. Sim, H. K. Kim, M. Y. Yoon, I. P. Hong, S. O. Woo, and Y.S. Choi, (2014), Detection and comparison of Nosema (Nosema spp.) in Apis mellifera and Apis cerana, Kor. J. Apicul, 29(4), p333-339.
[https://doi.org/10.17519/apiculture.2014.11.29.4.333]

- Fries, I., G. Ekbohm, and E. Villumstad, (1984), Nosema apis, sampling techniques and honey yield, J. Apicult. Res, 23, p102-105.
-
Fries, I., F. Feng, A. da Silva, S. B. Slemenda, and N. J. Pieniazek, (1996), Nosema ceranae n. sp. (Microspora, Nosematidae), morphological and molecular characterization of a microsporidian parasite of the Asian honeybee Apis cerana (Hymenoptera, Apidae), Eur. J. Protistol, 32, p356-365.
[https://doi.org/10.1016/S0932-4739(96)80059-9]

-
Higes, M., R. Martin, and A. Meana, (2006), Nosema ceranae, a new microsporidian parasite in honeybee in Europe, J. Invertebr. Pathol, 92, p93-95.
[https://doi.org/10.1016/j.jip.2006.02.005]

-
Higes, M., P. García-Palencia, R. Marín-Hernández, and A. Meana, (2007), Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia), J. Invertebr. Pathol, 94, p211-217.
[https://doi.org/10.1016/j.jip.2006.11.001]

-
Klee, J., W. T. Tay, and R. J. Paxton, (2006), Specific and sensitive detection of Nosema bombi (Microsporidia: Nosematidae) in bumble bees (Bombus spp.; Hymenoptera: Apidae) by PCR of partial rRNA gene sequences, J. Invertebr. Pathol, 91, p98-104.
[https://doi.org/10.1016/j.jip.2005.10.012]

-
Klee, J., A. M. Besana, E. Genersch, S. Gisder, A. Nanetti, D. Q. Tam, T. X. Chinh, F. Puerta, J. M. Ruz, P. Kryger, D. Message, F. Hatjina, S. Korpela, I. Fries, and R. J. Paxton, (2007), Widespread dispersal of the microsporidian Nosema ceranae, and emergent pathogen of the western honey bee, Apis mellifera, J. Invertebr. Pathol, 96, p1-10.
[https://doi.org/10.1016/j.jip.2007.02.014]

- Larsson, R., (1986.), Ultrastructure, function, and classification of Microsporidia, Prog. Protistol, 1, p325-390.
- Lee, B. R., J. N. No, P. V. Nguyen, M. S. Yoo, Y. H. Park, and B. S. Yoon, (2010), Development of Method for the Detection of Nosema ceranae by loop-mediated isothermal amplification, Kor. J. Apicul, 25(4), p267-274.
- Lim, H. Y., S. J. Yong, J. G. Lee, M. J. Ji, O. M. Lee, and B. S. Yoon, (2014), Development of specific PCR method for detection of Nosema apis based on nucleotide sequence, Kor. J. Apicul, 29(1), p27-33.
-
Nagamine, K., Hase, T., and Notomi, T., (2002), Accelerated reaction by loop-mediated isothermal amplification using loop primers, Molecular and Cellular Probes, 16, p223-229.
[https://doi.org/10.1006/mcpr.2002.0415]

-
Notomi, T., H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino, and T. Hase, (2000), Loop-mediated isothermal amplification of DNA, Nucleic Acids Research, 28(12), e63.
[https://doi.org/10.1093/nar/28.12.e63]

-
Smith, M. L., (2012), The honey bee parasite Nosema ceranae: transmissible via food exchange, PLOS ONE, www.plosone.org 7(8).
[https://doi.org/10.1371/journal.pone.0043319]

-
Ushikubo, H., (2004), Principle of LAMP method-a simple and rapid gene amplification method, Uirusu, 54(1), p107-112.
[https://doi.org/10.2222/jsv.54.107]

-
Wang, D.-I., and F. E. Moeller, (1970), The division of labor and queen attendance behavior of Nosema-infected worker honeybees, J. Econ. Entomol, 63, p1539-1541.
[https://doi.org/10.1093/jee/63.5.1539]

-
Webster, T. C., K. W. Pomper, G. hunt, E. M. Thacker, and S. C. Jones, (2004), Nosema apis infection worker and queen Apis mellifera, Apidologies, 35, p49-54.
[https://doi.org/10.1051/apido:2003063]

-
Weiss, L. M., C. R. Vossbrinck, (1999), Molecular biology, molecular phylogeny, and molecular diagnostic approaches to the microsporidia, In: Wittner, M., Weiss, L.M. (Eds.), The Microsporidia and Microsporidiosis, American Society for Microbiology, Washington, DC, p129-171.
[https://doi.org/10.1128/9781555818227.ch4]

-
Yamazaki, W., K. Seto, M. Taguchi, M. Ishibashi, and K. Inoue, (2008), Sensitive and rapid detection of cholera toxin-producing Vibrio cholerae using a loop-mediated isothermal amplification, BMC Microbiol, 8, p94.
[https://doi.org/10.1186/1471-2180-8-94]

-
Yamazaki, W., M. Taguchi, M. Ishibashi, M. Nukina, N. Misawa, and K. Inoue, (2009), Development of a loop-mediated isothermal amplification assay for sensitive and rapid detection of Campylobacter fetus, Veterinary Microbiology, 136, p393-396.
[https://doi.org/10.1016/j.vetmic.2008.11.018]

- Yoo, M. S., I. W. Kim, M. H. Kang, S. H. Han, and B. S. Yoon, (2008), Development of real-time PCR method for the detection of Nosema, Kor. J. Apicul, 23, p241-249.
- Yoo, M. S., P. V. Nguyen, J. N. No, B. R. Lee, Y. H. Park, and B. S. Yoon, (2011), Development of ultra-rapid real-time PCR method for the detection of Nosema, Kor. J. Apicul, 26, p21-27.
- Zander, E., (1907), Tierische Parasiten als Krankenheitserreger bei der Biene, Leipziger Bienenzeitung, 24, p164-166.


