
Antibacterial Peptide Gene Expression in the Honey Bee (Apis mellifera Feb.) Parasitized by Tropilaelaps Mite
Abstract
The mite, was originally confined to the wild honey bees in Asia, but now plagues the European honey bee, Apis mellifera. We amplified antimicrobial peptide cDNA genes (Defensin, Abaecin, Royalicin, and Hymenoptaecin) by RT-PCR in order to explore the transcriptional response to mite parasitism in A. mellifera, which differs in susceptibility to T. clareae, by comparing parasitized and non-parasitized full-sister adult bees (worker bees, drone bees) from the same hive. Differential gene expression in worker and drone bees induced by Tropilaelaps infestation was investigated by northern blotting. Tropilaelaps parasitism caused changes in the expression of genes related to sex distinction. Bees tolerant to viruses from Tropilaelaps were mainly characterized by differences in the expression of genes regulating antimicrobial gene expression. This finding provides a first step towards a better understanding of the molecular expression involved in this differential sex distinction host-parasite relationship. Therefore, this result demonstrated that Tropilaelaps were possible route of induction of antibacterial gene expression.
Keywords:
Antibacterial peptide, Apis mellifera, Abaecin, Hymenoptaecin, Defensin, Royalicin, Tropilaelaps, Bee virusINTRODUCTION
Honey bees are important economic eusocial insects that are vital for pollination and for the production of several bee products. The ectoparasitic Tropilaelaps mite feeds on the body fluids of the larvae, pupae, and adults of the western honey bee (Apis mellifera L.) causing serious damage to bee colonies. As eusocial insects, honeybees have evolved both communal and individual traits that reduce the impact of their numerous parasites and pathogens. Honeybees eliminate harmful invading pathogens and parasites through an inducible defense system (Boman et al., 1991) that includes at least four antimicrobial peptides (abaecin, defensin, apidaecin, and hymenoptaecin) (Casteels et al., 1989, 1993; Hoffmann et al., 1993). Following bacterial challenge, antibacterial peptides are produced in the fat body or hemocytes and then released into the hemolymph of insects. The following classes of insect antibacterial peptides have been proposed (Bulet et al.,1993; Casteels et al., 1993; Hultmark, 1993): (1) lysozymetype, (2) short α-helical, (3) short intra-disulfide linked, (4) short Arg-Pro-rich, and (5) long multi-domain. These antibacterial peptides function by deteriorating the bacterial cell envelope, with the exception of apidaecin, which is non-lytic (Casteels et al., 1994). In honeybees (Apis mellifera), the immune repertoire consists of four antibacterial peptides that are induced by bacterial infection and provide broad-spectrum antibacterial defense through complementarity (Casteels-Josson et al., 1994). Furthermore, induction of the entire antibacterial peptide repertoire in these bees can be triggered by infection with Escherichia coli alone (Casteels et al., 1989; Casteels-Josson et al., 1994). In the hemolymph of A. mellifera infected with E.coli, four different antibacterial peptides have been identified: apidaecin (Casteels et al., 1989), abaecin (Casteels et al., 1990), hymenoptaecin (Casteels et al., 1993) and defensin (Casteels-Jonsson et al., 1994). Recently, two structurally different defensin genes were cloned from A. mellifera (Klaudiny et al., 2005). It is believed that insects express a combination of antibacterial peptides in response to natural infection, leading to a broad spectrum of activity against microorganisms. Healthy animals can build up a variety of immune responses to microbial infections and septic wounds, and the rapid production of AMPs is one of the steps in this process.
AMPs are small bioactive molecules that are normally released in the hemolymph of infected invertebrate animals, such as honey bees. However, immune suppressed individuals will show greatly reduced responses. The AMP response is suppressed in varroa-parasitized bees, so that varroa infestation may suppress the bee immune system, resulting, for example, in increased titers of Deformed Wing Virus (DWV) and bees with visibly deformed wings. (Xiaolong and Cox-Foster, 2005).
We report here the detection of bee viruses and antibacterial gene products (abaecin, defensin, royalicin, and hymenopteacin) in A. mellifera based on sampling of adult worker and drone bees parasitized by T. clareae. We hypothesize that mites may suppress expression of genes that encode antimicrobial peptides. In addition, we have examined the differential expression between workers and drones. This research might help to explain how an ectoparasite can affect the immunity and pathology of its invertebrate host.
MATERIALS AND METHODS
Experimental animals
The honeybees (Apis mellifera) were collected from apparently infected colonies from an experimental apiary in Suwon and were analyzed using the RT-PCR for identification of virus and antibacterial genes. The live worker and drone honey bees were stored at -80°C until processing.
cDNA construction, genomic DNA isolation, and PCR
The whole bodies of A. mellifera worker, drone bees and Tropilaelaps mites were ground to a fine powder in liquid nitrogen for generation of cDNAs from mRNA. A 0.03g sample of powder was used to extract total RNA using a total RNA extraction kit (Promega, Madison, WI). RTPCR was performed using the Qiagen One Step RT-PCR kit (Qiagen, Germany) in 25μl reactions containing 200ng RNA templates and 2.5 pmol of each primer. Thermal reactions proceeded with an initial reverse transcription incubation at 42°C for 60 min then incubation at 95°C for 1 min. This was followed by 35 cycles of denaturation at 94°C for 30 s, annealing at 46°C for 45 s, extension at 72°C for 1 min. A final extension step was performed at 72°C for 10 min. PCR products were electrophoresed on a 1.2% agarose gel containing 0.5μg/ml ethidium bromide and subsequently visualized by UV transillumination. PCR products were excised using the Qiaex II gel extraction kit (Qiagen, Germany) following the manufacturer’s instructions and used as the templates for sequencing reactions. Genomic DNA was extracted from whole bodies of A. mellifera worker bees using the Wizard Genomic DNA Purification Kit (Promega, USA) and then used for Genomic PCR amplification with oligonucleotide primers designed from the cDNA sequences. Primers used were as follows: for abaecin, forward primer (5'-GATCTGCACACTCGAGGTCTG-3') and reverse primer (5'-GGGCGAATTGGGCCCGACTCGC-3'); for defensin, forward primer (5'-TGAGCTGTCGTTCAGACAC-3') and reverse primer (5'-TCGTTGAAT-CTTCATAATGGCAC-3'); for hymenoptaecin, forward primer (5'-TTCCAAGATGAAATTGATCGTGTTAG-3') and reverse primer (5'-TGCTACGTAAATACGAATCGTAATTTATTG-3'); for royalicin, forward primer (5'-CTTTGTGCTGGGAAGATGAAGTTC-3') and reverse primer (5'-GGGCGAATTGGGCCCGACGTCG-3'), based on the cDNA sequence. Genomic PCR was performed using 200ng RNA templates and 2.5 pmol of each primer. After 35 cycles of amplification (94°C for 30 s; 48°C for 40s; 72°C for 2 min), the PCR products were electrophoresed on a 1.2% agarose gel containing 0.5μg/ml ethidium bromide and subsequently visualized by UV transillumination. RT-PCR was performed using the Qiagen OneStep RT-PCR kit (Qiagen, Germany) in 25μl reactions containing 200ng RNA templates and 2.5 pmol each primer for detection of antibacterial peptides. Thermal reactions proceeded with an initial reverse transcription incubation at 50°C for 30 min then incubation at 95°C for 15 min. This was followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 45 s, extension at 72°C for 1 min. A final extension step was performed at 72°C for 10 min. PCR products were electrophoresed on a 2% agarose gel containing 0.5μg/ml ethidium bromide and subsequently visualized by UV transillumination.
DNA sequence analysis, genomic DNA isolation, PCR of antibacterial peptide genes
The RT-PCR (cDNA) and genomic PCR products were sequenced using an automatic sequencer (Model 3100 Genetic Analyzer; Perkin Elmer Applied Biosystems, Foster City, CA). Sequence data were automatically collated and analyzed using the ABI sequencing analysis software and subsequently manually verified. Similarities between virus sequences and published sequences were determined using BLAST (Basic Local Alignment Search Tool) within the National Centre for Biotechnology Information (NCBI) database (http://www.//ncbi.nlm.nih.gov/BLAST). Genomic DNA was extracted from fat body tissues of A. mellifera using the Wizard Genomic DNA Purification Kit (Promega, USA) and then used for PCR amplification with oligonucleotide primers designed from the cDNA sequences. After a 35-cycle amplification (94°C for 30 s; 48°C for 40 s; 72°C for 2 min), the PCR products were cloned into the pGem-T vector for sequencing (Promega, USA) and were transformed into E. coli TOP10F' cells (Invitrogen, Carlsbad, CA, USA). The nucleotide sequence was determined using a BigDye Terminator cycle sequencing kit and an automated DNA sequencer as described above.
RNA isolation and Northern blotting analysis
A. mellifera workers and drones were collected and washed twice with phosphate-buffered saline (PBS; 140mM NaCl, 27mM KCl, 8mM Na2HPO4, 1.5mM KH2PO4, pH 7.4). Total RNA was isolated from whole bee bodies using a Total RNA Extraction Kit (Promega, USA). Purified total RNA (1μg/lane) was separated on a 1.0% formaldehyde agarose gel, transferred onto a nylon blotting membrane (Schleicher & Schuell, Dassel, Germany), and hybridized at 46°C with a probe in hybridization buffer containing 5×SSC, 5×Denhardt’s solution, 0.5% SDS, and 100μg/ml denatured salmon sperm DNA. The cDNA PCR products were labeled with [α−32P]dCTP (Amersham, Arlington Heights, IL, USA) using a Prime-It Random Primer Labeling Kit (Stratagene, La Jolla, CA, USA) and used as a probe for hybridization. After hybridization, the membrane filter was washed three times for 30 min each in 0.1% SDS and 0.2×SSC at 65°C and then exposed to autoradiography film.
RESULTS AND DISCUSSION
Analysis antibacterial nucleotide sequencing and genomic DNA isolation and PCR
Four antibacterial peptide cDNAs (defensing, royalicin, hymenopteacin, abaecin,) were identified as having homology to previously reported antibacterial peptide genes. These cDNA clones, including the full-length open reading frames (ORFs), were sequenced and characterized (Fig. 1, 2, 3, and 4). We cloned A. mellifera defensin cDNA, a 398 bp long gene containing an ORF of 285 nuc-leotides capable of encoding a 95 amino acid polypeptide (Fig. 1A). The gene structure revealed that the A. mellifera defensin gene spans 2011 bp and contains two introns and three exons (Fig. 1B). From the sequence data, it appears that all insect defensins have a conserved region of six cysteine residues and share a common three-dimensional structure, made of an N-terminal loop, a central amphipathic α-helix, and a Cterminal antiparallel β-sheet (Bonmatin et al., 1992). In contrast to all other insect defensins, bee defensins have a 12 amino acid extension on their C-terminus, which encodes an additional C-terminal α-helix domain (Casteels-Josson et al., 1994). Moreover, the ORF of A. mellifera defensin has one extra amino acid (Gly) at the C-terminus, which may be amidated as suggested in the mature antibacterial peptide of A. mellifera defensin (Casteels-Josson et al., 1994). The structural similarity suggests that A. mellifera defensin has antibacterial activity, which has activity against Gram positive and Gram-negative bacteria (Rees et al., 1997).
And, we cloned A. mellifera royalicin cDNA, a 397 bp long gene containing an ORF of 309 nucleotides capable of encoding a 103 amino acid polypeptide (Fig. 2A). The gene structure revealed that the A. mellifera royalicin gene spans 1079 bp and contains two introns and three exons (Fig. 2B). The royalicin has very similar function of antibacterial activity with defencin.
We also cloned the A. mellifera hymenoptaecin cDNA, a 440 bp sequence that contains an ORF of 378 nucleotides that encodes a 126 amino acid polypeptide (Fig. 3A). The gene structure revealed that the A. mellifera hymenoptaecin gene spans 1968 bp and contains two intron and three exons (Fig. 3B). The reported antibacterial effects of A. mellifera hymenoptaecin suggest that it inhibits the viability of Gram negative and Gram-positive bacteria and its lethal effects against E. coli are secondary to sequential permeabilization of the outer and inner membranes (Casteels et al., 1993).
Finally, we also cloned A. mellifera abaecin cDNA according to results from searches of RT-PCR. The A. mellifera abecin cDNA is 354 bp long and contains an ORF of 157 nucleotides capable of encoding a 53 amino acid polypeptide. Comparisons of the amplicon size between the genomic DNA and cDNA sequences revealed that A. mellifera abaecin is an intron-less gene (Fig. 4). Comparative analysis suggests that the precursor molecule of A. mellifera abaecin is 53 amino acids long and contains only one possible processing site, resulting in a putative signal sequence of 19 amino acids (Casteels et al., 1990; Casteels-Josson et al., 1994; Rees et al., 1997). The 39-residue abaecin is a proline-rich antibacterial peptide with activity against both Gram-negative and Gram-positive bacteria (Rees et al., 1997).

cDNA sequence and structure of the Am-def gene. (A) Nucleotide sequence and predicated amino acid sequences of prepro-defencin cDNA clones numbers at the left indicate positions of the nucleotide and amino acid. The ATG start codon is boxed. The star symbol above the nucleotide sequence indicates the translational stop signal (TAA). (B) Genomic structure of the Am-def gene revealed by PCR amplification from Apis mellifera genomic DNA. Numbers indicate the position in the genomic sequences.
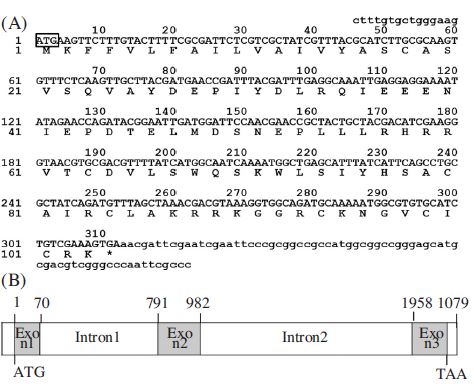
cDNA sequence and structure of the Am-royal gene. (A) Nucleotide sequence and predicated amino acid sequences of prepro-royalisin cDNA clones numbers at the left indicate positions of the nucleotide and amino acid. The ATG start codon is boxed. The star symbol above the nucleotide sequence indicates the translational stop signal (TGA). (B) Genomic structure of the Am-royal gene revealed by PCR amplification from Apis mellifera genomic DNA. Numbers indicate the position in the genomic sequences.

cDNA sequence and structure of the Am-hym gene. (A) Nucleotide sequence and predicated amino acid sequences of prepro-hymenoptaecin cDNA clones numbers at the left indicate positions of the nucleotide and amino acid. The ATG start codon is boxed. The star symbol above the nucleotide sequence indicates the translational stop signal (TAA). (B) Genomic structure of the Am-hym gene revealed by PCR amplification from Apis mellifera genomic DNA. Numbers indicate the position in the genomic sequences.

cDNA sequence and structure of the Am-abae gene. (A) Nucleotide sequence and predicated amino acid sequences of prepro-abaecin cDNA clones numbers at the left indicate positions of the nucleotide and amino acid. The ATG start codon is boxed. The star symbol above the nucleotide sequence indicates the translational stop signal (TAA).
Transcriptional expression profile of antibacterial peptide genes
The validity and quality of the PCR and northern hybridization results were further evaluated by examining the expression pattern of a selection of putative genes, based on their predicted involvement in mite infestation, by PCR and Northern hybridization (for detection of antimicrobial peptides). Differential expressions of four cDNAs from A. mellifera (for royalicin, defensin, abaecin, hymenopteacin) were compared between parasitized and normal bees (Fig. 5). The expressions of genes encoding four antimicrobial peptides (royalicin, defensin, abaecin and, hymenopteacin) were used as markers to measure the difference in the immune response between parasitized and normal bees (Fig. 3). We found that the transcriptional induction of three antibacterial peptide genes in the A. mellifera larvae significantly increased by Tropilaelaps mite. The transcriptional expression profile of antimicrobial peptide genes were expressed following sex specific induction or suppression by Tropilaelaps mites. The drone larva was not expression of abaecin, defensin and royalicin more than worker larva by Tropilaelaps mite infestations. However, hymenoptaecin is non up-regulated following by Tropilaelaps mite infestation compare with control (normal worker larva). Interestingly, hymenopteacin gene expression was suppressed in A. mellifera worker and drone by Tropilaelaps mite (Fig. 5(E)). We demonstrated that this ectoparasite could immunosuppress its invertebrate host with the evidence that parasitization significantly suppressed expression of some immunityrelated antibacterial genes. Ticks are known to immunosuppress their vertebrate hosts (Xiaolong and CoxFoster, 2005), so our finding indicates that immunosuppression of hosts may be a common antibacterial peptide in the interaction and coevolution between ectoparasites and their vertebrate and invertebrate hosts. The expression of the genes encoding the three antibacterial peptides was significantly upregulated by Tropilaelaps mite infestation. The worker bee was expressed antibacterial peptides more than drone bee. This work demonstrated that Tropilaelaps mite infestation suppressed hymenopteacin gene expression in honey bees. Not only the worker bees but also drone bees had suppressed hymenopteacin gene expression due to the ectoparasitization.
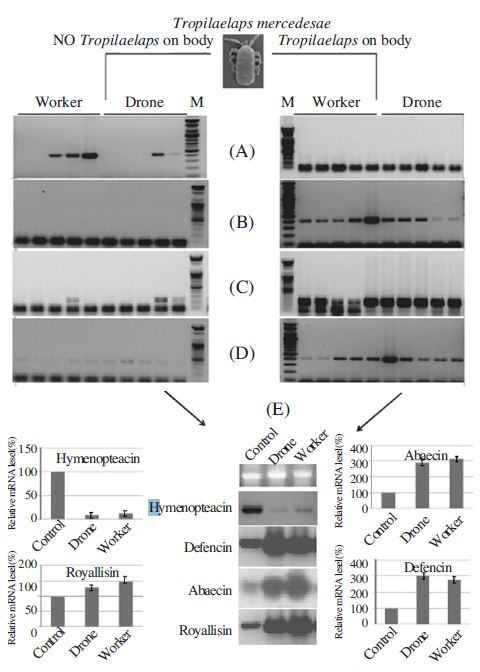
Sex distinction of the antimicrobial gene mRNA by RTPCR and Northern blot hybridization. Total RNA was isolated from the female and male bee, respectively. Induction of Abacin (A), Hymenopteacin (B), Defencin (C), and Royallisin (D) in female and male bee by and T. clareae. The template cDNA were diluted by nuclease free water. Transcriptional expression profile of antimicrobial peptide genes of sex specific induction by T. clareae (E). The RNA was separated by 1.0% formaldehyde agarose gel electrophoresis, transferred onto a nylon membrane, and hybridized with radiolabelled Abacin, Hymenopteacin, Defencin, and Royallisin cDNA. Ethidium bromide stain and Northern blot of rRNA are shown as a loading control.
Acknowledgments
This research was supported by an Agenda project (PJ01120102) from NAAS, Rural Development Administration.
LITERATURE CITED
-
Boman, H.G., Faye, I., Gudmundsson, G.H., Lee, J.Y., and Lidholm, D.A., (1991), Cell free immunity in Cecropia: a model system for antibacterial proteins, Eur. J. Biochem, 201, p23-31.
[https://doi.org/10.1007/978-3-642-77200-9_15]

-
Bonmatin, J.M., Bonnat, J.L., Gallet, X., Vovelle, F., Ptak, M., Reichhart, J.M., Hoffmann, J.A., Keppi, E., Legrain, M., Achstetter, T., (1992), Two-dimensional 1H-NMR study of recombinant insect defensin A in water: resonance assignments, secondary structure and global folding, J. Biomol. NMR, 2, p235-256.
[https://doi.org/10.1007/BF01875319]

- Bulet, P., Dimarcq, J.L., Hetru, C., Lagueux, M., Charlet, M., Hegy, G., Van Dorsselaer, A., Hoffmann, J.A., (1993), A novel inducible antibacterial peptide of Drosophila carries an O-glycosylated substitution, J. Biol. Chem, 268, p14893-14897.
- Casteels, P., Ampe, C., Jacobs, F., Vaeck, M., and Tempst, P., (1989), Apidaecins: antibacterial peptides from honeybees, EMBO J, 8, p2387-2391.
-
Casteels, P., Ampe, C., Riviere, L., Van Damme, J., Elicone, C., Fleming, M., Jacobs, F., Tempst, P., (1990), Isolation and characterization of abaecin, a major antibacterial response peptide in the honeybee (Apis mellifera), Eur. J. Biochem, 187, p381-386.
[https://doi.org/10.1111/j.1432-1033.1990.tb15315.x]

- Casteels, P., Ampe, C., Jacobs, F., and Tempst, P., (1993), Functional and chemical characterization of hymenoptaecin, an antibacterial polypeptide that is infectioninducible in the honeybee (Apis mellifera), J. Biol. Chem, 268, p7044-7054.
- Casteels, P., Romagnolo, J., Castle, M., Casteels-Josson, K., Erdjument-Bromage, H., and Tempst, P., (1994), Biodiversity of apidaecin-type peptide antibiotics, J. Biol. Chem, 269, p26107-26115.
- Casteels-Josson, K., Zhang, W., Capaci, T., Casteels, P., Tempst, P., (1994), Acute transcriptional response of the honeybee peptide-antibiotics gene repertoire and required post-translational conversion of the precursor structures, J. Biol. Chem, 269, p28569-28575.
-
Hoffmann, J.A., Hetru, C., Reichhart, J.M., (1993), The humoral antibacterial response of Drosophila, FEBS Lett, 325, p63-66.
[https://doi.org/10.1016/0014-5793(93)81414-U]

-
Hultmark, D., (1993), Immune reactions in Drosophila and other insects: a model for innate immunity, Trends Genet, 9, p178-183.
[https://doi.org/10.1016/0168-9525(93)90165-E]

-
Klaudiny, J., Albert, S., Bochanová, K., Kopernick´y, J., Simúth, J., (2005), Two structurally different defensin genes, one of them encoding a novel defensin isoform, are expressed in honeybee Apis mellifera, Insect Biochem. Mol. Biol, 35, p11-22.
[https://doi.org/10.1016/j.ibmb.2004.09.007]

-
Rees, J.A., Moniatte, M., Bulet, P., (1997), Novel antibacterial peptides isolated from a European bumblebee, Bombus pascuorum (Hymenoptera, Apoidea), Insect Biochem. Mol. Biol, 27, p413-422.
[https://doi.org/10.1016/S0965-1748(97)00013-1]

- Xiaolong, Y., and Cox-Foster, D.L., (2005), Impact of an ectoparasite on the immunity and pathology of an invertebrate: Evidence for host immunosuppression and viral amplification, PNAS 21, 21(102), p7470-7475.