Evaluation of Morphometric Characteristics to Discriminate Inbred Lines of the Honeybee Apis mellifera
Abstract
As a part of breeding project, five Apis mellifera inbreed lines (named line A, C, D, E and F) were bred and cultivated for several years via an artificial insemination method. In this study, we identified the 5 inbreed lines according to measured morphological values and conducted discriminant analysis. The morphometric characteristics analyzed include basitarsus length (ML), proboscis length (PL), fore wing length (FWL), Rs vein, distance (a), Rs vein, distance (b) and counted the number of hook (Ha). According to the this data, measured means of each body part showed significant differences by one-way ANOVA and post hoc Tukey HSD test (p<0.0001). To evaluate the individuals correctly classified, discriminant analysis was performed using 6 morphometric variables. When all characters were included, the analysis gave 77.4% separation of the individuals to their groups. Among them, line C was assigned 100% and line F was classified 90%, respectively, which determined more accurately classified than other groups. The result shows that the two inbreed lines controlled mating and presence of a more pure population.
Keywords:
Apis mellifera, Morphometric characteristics, Discriminant functionINTRODUCTION
Honeybees (A. mellifera) are generalist pollinators, thus important insects in agriculture, ecology and basic research(Jensen et al., 2005). Honeybee not only pollinator but also provide honey, royal jelly, propolis and pollen. In Korea, the economic value of honeybees as pollinators is reported to be more than over 29 billion dollars (Jung, 2008) and the demand for pollination also has increased drastically. However, honeybees face challenges, including pesticides, predators (such as hornets), pathogens and parasites (such as mites) have contributed to their decline (Bakonyi et al., 2002; Bailey and Ball, 1991; Ingram et al., 1996; Oldroyd, 2007).
Honeybee queens are polyandrous (Laidlaw and Page, 1985; Adams et al., 1977), meaning mating with two or more males and this behaviour contributes higher genetic diversity. Genetic diversity directly influences on colony productivity, disease and pest resistance to parasites and pathogens and resultantly enhances colony fitness via an enhanced task allocation system (Oldroyd and Fewell, 2008). Therefore, the loss of genetic diversity is of grave concern. It has been shown that colonies with reduced genetic diversity are less capable of controlling hive temperature and more prone to develop diseases when challenged by parasites (Tarpy, 2003). From the breeding perspective view, however, this behaviour makes selection of desired characters difficult, because several male traits are inherited simultaneously to the offspring in a colony. Moreover, honeybees are particularly sensitive to inbreeding (Seeley and Tarpy, 2007). Thus, there is a widely recognized need to encourage regional breeding efforts to preserve local adaptation and to maintain local strains in isolated conservation apiaries.
Morphometric characteristics of honeybees have also been well studied for the identification of geographic races or subspecies, based on multiple measurements of several individuals (Alpatov, 1929). Ruttner et al. (1978) chose 42 characteristics for analysis of honeybee workers from a wide range of geographic locations. Since then, more accurate bases for determining geographical races of bees using biometrics have been proposed by other researchers (Ruttner, 1988; Ruttner et al., 2000; Radloff et al., 2003; Diniz-Filho et al., 2000). The standard morphometric characteristics of honeybees include distances of body parts, venation angles and discrete classes of pigmentation. Recently, some researchers have been conducted discriminant analysis based on the morphometric characteristics. Discriminant analysis is a technique in which measurements of two or more characters are weig-hted and combined linearly to give maximal separation of two or more groups (Blackith and Reyment, 1971; Van de Geer, 1971). DuPraw (1964, 1965a, 1965b) first applied discriminant analysis to measure features of the wings and after that many researchers has been conducted to classifying samples of various subspecies led to other multivariate applications.
In a previous study, we have been conducting a breeding project for Apis mellifera at the National Academy of Agricultural Science (NAAC) in Korea (Kim et al., 2015). According to the data, the 6 inbred lines have been maintained under the controlled production of queens and artificial insemination. In this study, we evaluated morphometric characteristics based on different body parts and conducted discriminant analysis in 5 inbred lines, A, C, D, E and F.
MATERIALS AND METHODS
Control mating by instrumental insemination
The honey bee breeding line has been created through the production of queen from selected colony and controlled mating was carried out by artificial insemination over 8 years (Kim et al., 2015). There were 5 lines named A, C, D, E and F. Each line had 3 colonies that were kept in standard Langstroth hives. Breeding lines were instrumentally inseminated from selected colonies of each line or cross breeding scheme. Queens and drones of each line were reared in favorable floral condition at National Institute of Agricultural Sciences (NIAS) in Korea. Queen rearing was done while drones were started emerging. Virgin queens were kept in nucleus of 8 frames standard Langstroth hives. When the virgin queen was 6 days post emergence, she was anesthetizing treated with 5 minutes dose of carbon dioxide prior artificial insemination date one day. At the date of insemination, semen of selected drones was collected into glass capillary of Harbo syringe at the age about 2 weeks after they emerged. Virgin queen was narcotized on queen holder in Schley equipment model to manually open sting chamber and inject semen to vagina. One virgin queen was instrumentally inseminated with a dose of 8μl semen. After insemination, one wing of queen was one third clipped and she was marked with assigned number on thorax.
Investigation of morphometric characteristics
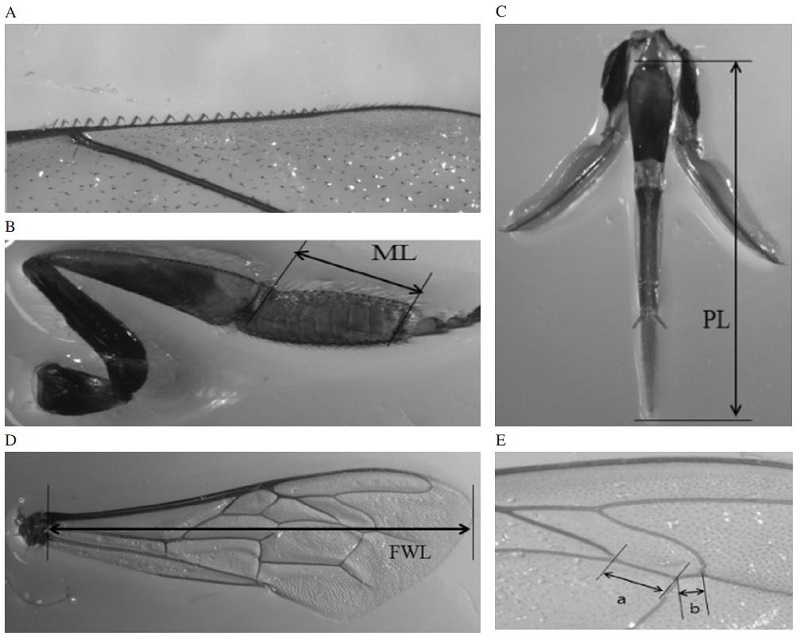
The standard morphometric characteristics were analyzed in 10 workers bees of each of 3 colonies of 5 inbreed lines, A, C, D, E and F. These characteristics are described in Ruttner (1988) and Alpatov (1929), and include the following: number of the hooks (Ha), basitarsus length (ML), proboscis length (PL), forewing length (FWL), Rs vein, distance (a), and Rs vein, distance (b). Fig. 1 shows standard morphometric characteristics of honeybees. Adult workers from 15 colonies were sampled from an apiary in NAAS and 10 workers were usually measured from each colony and constitute a collection in this study. Honeybees were preserved in 95% ethanol, and were then dissected and subsequently mounted in diaphane or permount on a slide and measured using Leica DM2500 (Leica microsystems, Germany) microscope. Wing venation and size characteristics were analyzed using a microscope and Leica Application Suite (LAS) V4.7 software.
Statistical analysis
To compare the character of morphological, the statistical analysis was conducted using one-way analysis of variance (ANOVAs PROC ANOVA; SAS Institute, 1989) among the five inbreed lines. When Anova result showed the significant difference between the mean of treatment, the multiple range test using Tukey’s was conducted to differentiate the significant difference between the mean of treatment. The discriminant analysis was performed using SPSS v.15.
RESULT
Measurement of morphometric characteristics
Table 1 and Fig. 2 show measured morphometric characteristics in this study. The five inbreed lines exhibited statistically significant differences in fore wing length (FWL), varying from 8.2 to 9.1mm (one-way ANOVA and post hoc Tukey’s HSD test (p<0.05)). The highest fore wing length was observed in line D (9.1mm, N=35) and the lowest was observed in line C (8.2mm, N=26). The average basitarsus length (ML) in the five inbreed lines examined ranged from 1.7 to 2.0mm (p<0.05). The highest basitarsus length was found same for line A (2.0mm, N=37), D (2.0mm, N=35) and F (2.0mm, N=35), and the lowest in line C (1.7mm, N=26). The number of hooks (Ha) ranged from 20.6 to 22.5 (p<0.05). The highest number of hooks observed in line C (22.5mm, N=26) and the lowest was line A (20.6mm, N=37). The mean proboscis length (PL) showed no significant differences (p>0.05) among the 5 inbred lines, and it ranged from 5.3 to 5.7mm. The highest length of proboscis was observed in line A (5.7, N=37) and D (5.7mm, N=35) and the lowest was line C (5.3mm, N=26). The mean rs vein, distance (a) showed no significant differences (p>0.05) among the 5 inbred lines, ranging from 1.98 (F, N=35) to 2.87mm (D, N=35). However, the mean of rs vein, distance (b) showed significant differences (p<0.05). The ratio of rs vein distances a and b ranged from 1.98 (F, N=35) to 2.87mm (D, N=35).
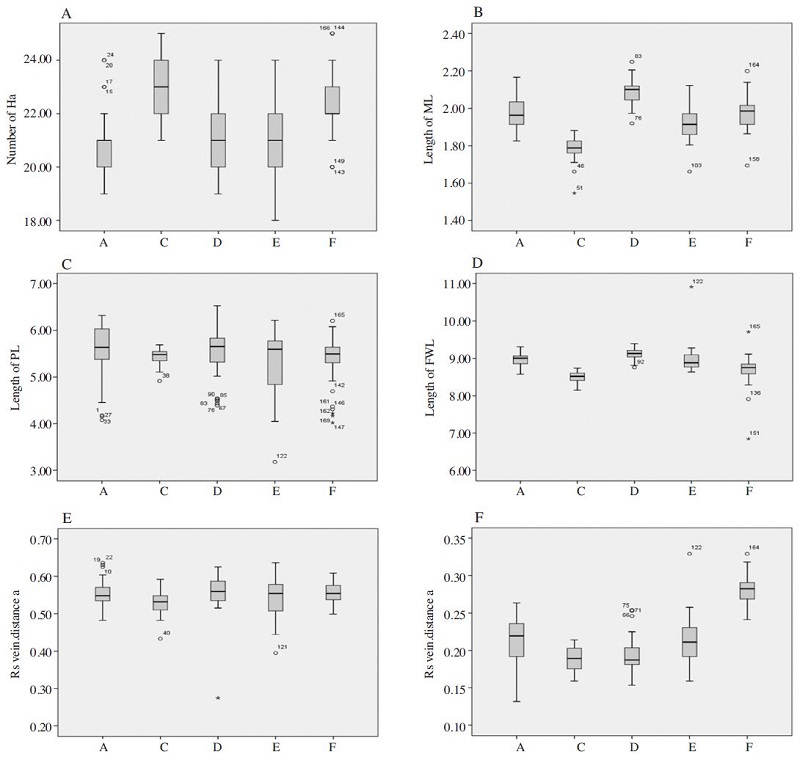
Measured morphometric characteristics from different body parts. (A); Number of the hooks (Ha), (B); Measurement of the basitarsus length (ML), (C); Measurement of the proboscis length (PL), (D); Measurement of the fore wing length (FWL), (E); Rs vein (distance a in Fig. 3) and (F); Rs vein (distance b in Fig. 3). The boxplot diagram shows the median (the line in the box), means (+), interquartile range and outliers (○, *).
Discriminant function analysis
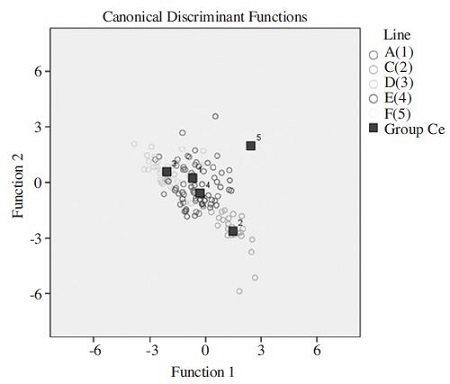
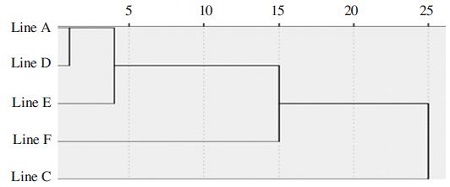
A discriminant function analysis of the five inbreed lines carried out on the data analyzed morphometric characteristics showed that the characteristics evaluated statistically significant differences among individuals of 5 inbred lines. The result of the discriminant function analysis of individuals indicated that the 5 inbred lines accurately classified 77.4% in their original groups. Among the 164 honeybees, 127 honey bees were assigned to their actual groups, whereas, 37 honey bees were scattered (Table 2). Specifically, line C was assigned to actual groups by 100% and line F was classified F line by 90%, but line A, D and E were assigned to actual groups by 45.9%, 88.9% and 71.4%, respectively. According to mahalanobis distance, the centroids of lines A, D and E showed partial overlapping, which found to be morphologically the closest. However, the mahalanobis distance between the centroids of lines C and F was separated, which mean the two lines morphologically difference (Fig. 3). The result of that five inbreed lines based on the six morphological characteristics was found separated for 3 groups (P<0.001). The relationship among 5 inbred lines is shown with the help of a dendrogram (Fig. 4). According to data from cluster analysis, the lines A, D, and E clustered closely together in the dendrogram.
DISCUSSION
As part of a breeding project, five Apis mellifera inbreed lines (named lines A, C, D, E and F) were bred and cultivated for several years (Kim et al., 2015). In this study, we discriminated maintained 5 inbreed lines based on standard morphometric characteristics. According to the results, the means of the measurements of each body part showed considerable differences among the 5 inbred lines. Of these, significant differences were found in fore wing length, hook number, rs vein, distance (b), and basitarsus length. Discriminant function analysis resulted in the clear separation of 5 inbred lines when 6 morphometric variables was utilized (Fig. 3). Among them line C was line F was classified to actual groups by 100% and 90%, respectively, which determined more accurately classified than other groups. Discriminant function analysis of 5 inbred lines was also supported by the Dendrogram based on Mahalanobis distances (Fig. 4). The C and F lines separated from other lines. The result shows that the two inbreed lines controlled mating and presence of a more pure population. In general, the standard morphometric characteristics that are used to discriminate honeybee species and subspecies. However, the discriminance among same subspecies are difficult by analyzing only a few morphological characteristics. But, in this study, the validated characteristics showed differences among the 5 lines, especially, line C and F showed morphologically distinguishable.
Acknowledgments
This study was carried out with the support of "Next-Generation BioGreen 21 Program (PJ01117001)", Rural Development Administration, Republic of Korea.
References
- Adams, J., Rothman, E.D., Kerr, W.E., and Z.L. Paulino, (1977), Estimation of the number of sex alleles and queen matings from diploid male frequencies in a population of Apis mellifera, Genetics, 86, p583-596.
-
Alpatov, W.W., (1929), Biometrical studies on variation and races of the honeybee (Apis mellifera L.), The Quarterly Review of Biology, 4(1), p1-58.
[https://doi.org/10.1086/394322]

- Bailey, L., and B.V. Ball, (1991), Honey Bee Pathology, second ed, Academic Press, London, UK.
-
Bakonyi, T., Farkas, R., Szendroi, A., Dobos-Kovacs, M., and M. Rusvai, (2002), Detection of acute bee paralysis virus by RT-PCR in honey bee and Varroa destructor field samples: rapid screening of representative Hungarian apiaries, Apidologie, 33, p63-74.
[https://doi.org/10.1051/apido:2001004]

- Blackith, R.E., and R.A. Reyment, (1971), Multivariate morphometrics, Academic Press, London, p412.
-
Diniz-Filho, J.A.F., Hepburn, H.R., Radloff, S., and S. Fuchs, (2000), Spatial analysis of morphological variationnin African honeybees (Apis mellifera L.) on a continental scale, Apidologie, 31, p191-204.
[https://doi.org/10.1051/apido:2000116]

-
DuPraw, E.J., (1964), Non-Linnean taxonomy, Nature, 202, p849-852.
[https://doi.org/10.1038/202849a0]

-
DuPraw, E.J., (1965a), Non-Linnean taxonomy and the systematics of honeybees, Syst. Zool, 14, p1-24.
[https://doi.org/10.2307/2411899]

- DuPraw, E.J., (1965b), The recognition and handling of honeybee specimens in non-Linnean taxonomy, J. Apicult. Res, 4, p71-84.
-
Harbo, J.R., (1986), Propagation and instrumental insemination. Bee genetics and breeding, Academic Press, Orlando, Fla, p361-389.
[https://doi.org/10.1016/B978-0-12-588920-9.50020-0]

- Ingram, M., Nabhan, G.C., and S.L. Buchmann, (1996), Impending pollination crisis threatens biodiversity and agriculture, Tropinet, 7, p1.
-
Jensen, A.B., Palmer, K.A., Boomsma, J.J., and B.V. Pedersen, (2005), Varying degrees of Apis mellifera ligustica introgression in protected populations of the black honeybee, Apis mellifera mellifera, in northwest Europe, Molecular Ecology, 14, p93-106.
[https://doi.org/10.1111/j.1365-294X.2004.02399.x]

- Jung, C.E., (2008), Economic Value of Honeybee Pollination on Major Fruit and Vegetable Crops in Korea, Korean Journal of Apiculture, 23(2), p147-152.
-
Kim, H.K., I. Kim, M.L. Lee, Y.S. Choi, and B.R. Jin, (2015), Microsatellite markers developed by next-generation sequencing differentiate inbred lines of Apis mellifera, Journal of Asia-Pacific Entomology, 18, p801-805.
[https://doi.org/10.1016/j.aspen.2015.10.003]

- Laidlaw, H.H., and R.E. Page, (1985), Polyandry in honeybees (Apis mellifera L.): sperm utilization and intracolony genetic relationships, Genetics, 108, p985-997.
- Laidlaw, H.H., and R.E. Page, (1997), Queen rearing and bee breeding, Cheshire, Conneticut, USA, Wicwas Press.
-
Oldroyd, B.P., (2007), What's killing American honey bees?, PLoS Biol, 5, pe168.
[https://doi.org/10.1371/journal.pbio.0050168]

-
Oldroyd, B.P., and J.H. Fewell, (2008), Large fitness benefits from polyandry in the honeybee, Apis mellifera, Trends in Ecology & Evolution, 23, p59-60.
[https://doi.org/10.1016/j.tree.2007.10.012]

-
Radloff, S.E., Hepburn, R., and L.J. Bangay, (2003), Quantitative analysis of intracolonial and intercolonial morphometric variance in honeybees, Apis mellifera and Apis cerana, Apidologie, 34, p339-351.
[https://doi.org/10.1051/apido:2003034]

-
Ruttner, F., (1988), Biogeography and taxonomy of honeybees, Springer, Berlin.
[https://doi.org/10.1007/978-3-642-72649-1]

-
Ruttner, F., Elmi, M.P., and S. Fuchs, (2000), Ecoclines in the near east along 36 degrees N latitude in Apis mellifera L, Apidologie, 31, p157-165.
[https://doi.org/10.1051/apido:2000113]

-
Ruttner, F., Tassencourt, L., and J. Louveaux, (1978), Biometrical statistical analysis of the geographic variability of Apis mellifera L, Apidologie, 9, p363-381.
[https://doi.org/10.1051/apido:19780408]

-
Seeley, T.D., and D.R. Tarpy, (2007), Queen promiscuity lowers disease within honeybee colonies, Proceedings of the Royal Society B-Biological Sciences, 274, p67-72.
[https://doi.org/10.1098/rspb.2006.3702]

-
Tarpy, D.R., (2003), Genetic diversity within honeybee colonies prevents severe infections and promotes colony growth, Proceedings of the Royal Society of London, B, 270, p99-103.
[https://doi.org/10.1098/rspb.2002.2199]

- Van de Geer, J.P., (1971), Introduction to multivariate analysis for the social sciences, W. H. Freeman and Company, San Francisco, p293.