
Metabolomic Studies in Apis mellifera
Abstract
The application of metabolomic analysis has emerged as a potent tool for exploring honeybee physiology. This technique enables researchers to detect thousands of chemical compounds simultaneously in a single sample, which represent a combination of gene expression, enzyme activity, and environmental factors. The utilization of metabolomics has facilitated a broad range of investigations in honeybees, yielding novel insights into gene function and metabolic status that would otherwise be unattainable. The present review outlines the applications of metabolomic analysis since its introduction in the field, while also discussing the main techniques employed for metabolomic studies in honeybees. Furthermore, we identify and highlight the new avenues for future research in this field.
Keywords:
Metabolomics, Honeybees, Pathogens, Pesticides, DietsINTRODUCTION
Pollinators are vital for maintaining ecosystem equilibrium, enabling sustainable agriculture, and ensuring food security (Potts et al., 2016). Animal pollination is necessary for the production of roughly 75% of vegetable food crops and seeds, which increases crop quality, shelf life, and commercial value (Klein et al., 2007). Despite being the most critical pollinators worldwide, Apis mellifera honeybee colonies have declined globally in recent decades, attracting considerable attention (vanEngelsdorp and Meixner, 2010). Climate change, habitat fragmentation and loss, invasive species, pathogens, and agrochemicals are suspected of negatively impacting their survival (Higes et al., 2007; Genersch, 2010; Henry et al., 2012; Guez, 2013). Agrochemicals are extensively utilized in agriculture and pose a significant threat to populations, as they can easily expose and impact them during foraging and pollination, causing widespread concern.
To elucidate disorders and timely detect alterations in bee physiology, it is crucial to employ molecular instruments for screening the gamut from genome to metabolome (Lankadurai et al., 2013). Of these instruments, metabolomics is the most informative, as it focuses on the final stage of the “omics cascade” by examining small metabolites and strongly correlates with the phenotype (Alonso et al., 2015; Liu and Locasale, 2017). After the completion of whole genome sequencing of various insect species such as fruit flies, mosquitoes, and honeybees, research and development in the field of omics, including transcriptomics, have been actively pursued to understand the functions of individual genes. Among them, metabolomics is a research field that tracks quantitative or qualitative changes in metabolites within an organism and interprets various physiological phenomena or mechanisms at the cellular or organismal level in a comprehensive manner based on this information.
Insect metabolomics research, using flies and mosquitoes, is actively ongoing, and research on honeybee metabolism has greatly increased since the 2010s (Table 1). Honeybee metabolomics research, specifically, is a study that comprehensively understands the complex physiological phenomena of honeybees by tracking and investigating the temporal and spatial changes of all metabolites present in honeybee tissues in response to various internal and external environmental conditions, such as stress responses to behavior, infection, and temperature changes, as well as symbiosis with bacteria (Chakrabarti et al., 2019; Lee et al., 2022; Ricigliano et al., 2022; Kunc et al., 2023). Currently, the field of honeybee metabolomics research is widely used as a research tool for functional genomics, including the elucidation of metabolic regulatory mechanisms and the understanding of physiological responses, due to the biological information provided by previously known individual honeybee genomics and the development of analytical instruments.
The goal of metabolism research is to understand and interpret the metabolic processes and pathways within cells and tissues. Therefore, honeybee metabolism research is also evolving in the direction of understanding metabolic pathways from a systems biology perspective for the purpose of identifying various unknown metabolic pathways (Fig. 1). In this paper, we aim to introduce the current status of honeybee metabolism research.

A descriptive overview is given regarding the procedure of extracting and studying metabolites. Honeybee metabolomic studies were conducted under four different conditions, including nutrition, climate change, pesticide exposure, and microbiome analysis. Samples were collected from bee larvae, adult bees, and specific tissues, and their metabolic compositions were examined using untargeted and targeted liquid chromatography-mass spectrometry (LC-MS) or gas chromatography-mass spectrometry (GC-MS) techniques. The data obtained from LC-MS or GC-MS were processed using specialized metabolomics software.
METHODS USED FOR METABOLOMIC ANALYSIS IN HONEYBEES
The metabolome refers to any small molecule (<1,500 Da) that an organism ingests, produces, catabolizes, or encounters in its surroundings (Wishart et al., 2007). While the specific composition of the honeybee metabolome has not yet been defined, the Human Metabolome Database currently contains information about more than 42,000 small molecules (Wishart et al., 2013). This number is expected to increase as technology advances. Given the high conservation of metabolic pathways between flies and humans (Rajan and Perrimon, 2013), it is likely that the honeybees metabolome also encompasses a similar range of compounds. However, due to the diverse nature of these molecules, no single method can effectively measure all of them. Consequently, each study must focus on measuring a specific subset of the metabolome.
The majority of metabolomic investigations rely on three main types of equipment: Gas Chromatography/Mass Spectrometry (GC-MS), Liquid Chromatography/Mass Spectrometry (LC-MS), and Nuclear Magnetic Resonance (NMR). Each of these methods has its own advantages and disadvantages, and although the availability of equipment often determines the choice of analysis, comprehending the capabilities of each instrument can greatly enhance the efficiency of study design.
APPLICATIONS OF METABOLOMICS IN APIS MELLIFERA
Despite the fact that the technology needed for metabolomic research has only been accessible for about 15 years, more than 20 studies in honeybees have taken advantage of this technology, encompassing a broad spectrum of topics as shown in Table 1. These applications have demonstrated their significance in the identification of metabolites that are specific to certain tissues, and in examining the impacts of pathogens, diets, environmental conditions, or pesticide stresses on the organism’s overall physiology, also demonstrated in Table 1. This type of analysis has frequently provided unique insights into metabolism that would not have been possible using any other methods.
1. Comparative analysis of metabolites among honeybee groups according to season (summer and winter)
Due to global warming, climate change has occurred, leading to a decrease in the habitat of honeybees and a subsequent decrease in the number of honeybee populations (Le Conte and Navajas, 2008). The gradual and consistent reduction in the capacity of honeybee populations to survive through winter in different parts of the world, such as North America and Europe, is a concerning occurrence that has been reported in various studies (vanEngelsdorp et al., 2015; Gray et al., 2019). In the last few decades, an increase in mortality rates during overwintering has been observed (vanEngelsdorp et al., 2009), which is likely attributable to a combination of biotic and abiotic stressors. These stressors include the Varroa destructor mite (van Dooremalen et al., 2012), Nosema sp. Microsporidia (Higes et al., 2008), viral infections (Highfield et al., 2009), exposure to pesticides (Simon-Delso et al., 2014), dietary and nutritional deficiencies (Requier et al., 2017), unfavorable winter conditions (Steinhauer et al., 2015), and climate change (Le Conte and Navajas, 2008). Depending on the temperature change, the honeybee population can be divided into two groups: summer bees with a short lifespan and winter honeybees with a long lifespan. Summer bees mainly engage in brood rearing, nectar and pollen collection, and have a lifespan of about 15-38 days. On the other hand, as brood rearing begins to decline in the fall, winter bees begin to emerge, and their main task is to keep the colony warm throughout the winter and to raise the first new generation in late winter or early spring, with a lifespan of about 140-320 days (Page and Peng, 2001).
Although many studies have revealed the biochemical parameters of the difference in lifespan between summer honeybees and winter honeybees, relatively few studies have been conducted on metabolic changes within honeybees. Recently, differences in metabolic profiling between summer honeybees and winter honeybees were revealed through 1H NMR analysis (Lee et al., 2022). According to the results, there were 28 metabolites that showed statistically significant differences between the two groups, which can be divided into carbohydrates, amino acids, choline-containing compounds, and unknown compounds based on the metabolite groups. Compared to summer bees, winter bees had higher concentrations of fructose, sucrose, and trehalose, while their levels of amino acids, except for proline and alanine, were lower than those of summer bees. Carbohydrates such as sucrose and trehalose are essential energy sources for bees and are stored as glycogen in the fat body (Blatt and Roces, 2001). Therefore, high levels of carbohydrates can be converted to high levels of glycogen, which can be used as a survival energy source during the winter. Additionally, trehalose is known to protect bees from various stresses such as dryness, low temperature, and hibernation (Watanabe et al., 2002; Kostal et al., 2007; Overgaard et al., 2007). Therefore, the high trehalose levels in winter bees are considered an adaptive response to withstand cold temperatures and other environmental stresses.
Proline is the primary amino acid found in the hemolymph of honeybees (Hrassnigg et al., 2003) and is recognized as an insect cryoprotectant, similar to trehalose (Kostal et al., 2012). Studies on freeze tolerance in fruit flies (Drosophila melanogaster) have shown that proline and trehalose are the most abundant metabolites accumulated during cold adaptation (Kostal et al., 2012; Williams et al., 2016). Therefore, the elevated levels of trehalose and proline in winter bees function as cryoprotectants, safeguarding the integrity of cellular membranes during the cold winter period, maintaining optimal water levels, and serving as an energy reservoir.
Meanwhile, Lee et al. (2022) showed results that adenosine monophosphate (AMP) and oxidized nicotinamide adenine dinucleotide (NAD+), which are nucleotide metabolites, were also produced more highly in winter bees than in summer bees. The changes in the concentrations of AMP and NAD+ are closely related to the cellular energy metabolism, and it is known that they regulate the expression of AMP-activated protein kinase (AMPK) and sirtuin genes (Hardie, 2007; Bonkowski and Sinclair, 2016), thereby regulating lifespan. In other words, activated AMP and NAD+ have been shown in studies using C. elegans and D. melanogaster to activate the expression of AMPK and sirtuin, thereby extending lifespan. Therefore, the increased expression of AMP and NAD+ in winter bees can be considered as one of the switches that promote the transition from a short-lived generation (summer bees) to a long-lived generation (winter bees).
2. Metabolic profiles of honeybees in response to changes in artificial flower composition
When nutrient sources are limited, the productivity of honeybees is negatively impacted. Inadequate nutrition causes a decrease in the supply of replacement bees, leading to a decline in the number and vitality of bees within colonies, as well as a reduction in their resistance to prevalent diseases (Paray et al., 2021). Therefore, managed honeybee colonies are commonly provided with artificial diets known as “pollen substitutes” due to insufficient pollen availability in the environment and the consequent risk of malnutrition. Protein-rich components such as soy, corn gluten are typically used in various diet formulations to supply essential amino acids (Brodschneider and Crailsheim, 2010). However, despite the widespread use of these diets, their effectiveness compared to natural pollen remains uncertain. Recent studies have explored the potential of microalgae as a dietary supplement for managed honeybees (Madeira et al., 2017). Notably, Chlorella, a eukaryotic microalga, and Arthrospira, a prokaryotic cyanobacterium commonly known as spirulina, have been identified as rich sources of protein, fatty acids, sterols, and other bioactive compounds with nutraceutical properties. These microalgae are easily digestible and have been found to mimic the growth characteristics of a natural pollen diet. However, despite promising results, the underlying metabolic mechanisms responsible for the positive effects of microalgae on bee health remain poorly understood.
Ricigliano et al. (2022) have found that microalgae can be used as a feed additive, which can be utilized as a natural product to improve honeybee health. They added Chlorella vulgaris and Arthrospira platensis to the feed and provided it to the honeybees to study the changes in feed consumption and metabolite profiles. Untargeted metabolite analysis was performed using LC-MS and GC-MS to measure the metabolite profiles of honeybees that consumed the natural feed and those that consumed the feed containing microalgae. Two groups of honeybees that consumed different diets shared 248 metabolites in LC-MS and 87 metabolites in GC-MS. Twenty-five specific metabolites were identified according to the diet, which were found to contain lipids, essential fatty acids, vitamins, and plant compounds. The honeybees that consumed pollen diet with algae showed higher levels of linoleic acid, zeaxanthin, lutein, α-tocopherol, and β-carotene. These metabolite analysis results confirmed that algae such as Chlorella included in artificial pollen diet can regulate honeybee growth, antioxidant activity, and heat shock protein gene expression. Therefore, algae like Chlorella can be used as sustainable artificial supplementary and can be utilized as a natural product to improve honeybee health. This metabolite analysis provides important information for the development of precise nutritional management of honeybee diet.
3. Comparative analysis of honeybee metabolites in response to insecticide exposure
Pesticides have a prevalent application in agriculture, albeit posing significant threats to the well-being of numerous non-target organisms (Desneux et al., 2007; Schafer et al., 2012). In recent times, there has been a significant focus on the impact of neonicotinoid pesticides and their potential to harm the health of honeybee colonies worldwide (Blacquiere et al., 2012). Neonicotinoid insecticides act as systemic insecticides, causing abnormal excitement and convulsive paralysis in insects, and are widely used in agriculture to control various types of pests (Brown et al., 2006; Simon-Delso et al., 2015). The exposure to insecticides is not limited to direct contact; rather, it can also occur through the ingestion of contaminated food sources, such as nectar, pollen, and water. Extensive research has reported the presence of neonicotinoid insecticides such as imidacloprid, thiamethoxam, and thiacloprid residues in various bee-derived products, including honey, beeswax, and pollen, as documented in several scientific studies (Codling et al., 2016; Sanchez-Hernandez et al., 2016; Tosi et al., 2018). Most neonicotinoid insecticides are highly toxic to bees and can induce excessive toxicity reactions, but it has been found that outdoor insecticide doses do not cause acute toxicity (Blacquiere et al., 2012). However, research reports have been published indicating that exposure to sublethal doses of neonicotinoid insecticides can affect brain and thorax development (Oliveira et al., 2014), impair learning and memory (Aliouane et al., 2009), and destroy the immune system in bees (Brandt et al., 2016). Furthermore, many outdoor experiments have shown that neonicotinoid insecticides can reduce the foraging and cognitive abilities of bees returning to their hives (Fischer et al., 2014). In a recent study, an investigation was conducted using LC-MS and GC-MS-based metabolomic techniques to explore the metabolic profile variations between honeybees treated with thiacloprid and those in the control group (Shi et al., 2018). The aim of this investigation was to gain a better understanding of the intricate interactions between neonicotinoids and honeybees at the molecular level. Researchers investigated the metabolite profiles of bee heads exposed to 2 mg/I of thiacloprid continuously for three days using both LC-MS and GC-MS. The analysis showed that 115 metabolites showed differences in expression compared to the control group, with 80 metabolites appearing in the LC-MS analysis and 35 metabolites appearing in the GC-MS analysis. The 115 metabolites included various amino acids, fatty acids, phospholipids, and other compounds, with over 70% of these metabolites showing higher expression than in the control group. Interestingly, among the metabolites that showed differences in expression, thiacloprid itself or its metabolites were not detected. This suggests that thiacloprid is rapidly metabolized in the thorax after being absorbed by bees, so only minimal amounts of thiacloprid can reach the bee head. According to previously published research results, when detoxification of alkaloid nicotine occurs in bees, metabolic activity, antioxidant and heat shock responses are increased (Rand et al., 2015). Genes induced by the neonicotinoid thiamethoxam that show differences in expression are primarily abundant in biosynthetic and metabolic pathways related to ribosomes, tyrosine metabolism, and drug metabolism pathways.
4. Effects of pathogen infection with Nosema cerana
Nosema ceranae, a gut parasite prevalent in A. mellifera, is widely distributed worldwide and poses a significant threat to the Western honeybee, both at the individual and colony levels (Goulson et al., 2015). As with other microsporidian species, N. ceranae is capable of manipulating bee physiology and behavior to create a more favorable environment for its reproduction. N. ceranae infection has been shown to weaken the integrity of the midgut tissue (Dussaubat et al., 2012), alter energy demand (Alaux et al., 2009) and hemolymph sugar levels (Mayack and Naug, 2010), and suppress the immune response of honeybees (Alaux et al., 2009). Infected honeybees have been found to have shorter lifespans than uninfected honeybees in some studies, but the presence of N. ceranae is not always associated with honeybee mortality, indicating variations in the parasite’s virulence (Vidau et al., 2011). These variations may be influenced by host or parasite genetics, climate, nutrition, or interactions with other stressors such as environmental contaminants or other parasites.
Jousse et al. (2002) have developed a novel approach for conducting metabolomics analysis on hemolymph samples through a combination of LC/MS and NMR techniques. Their objective was to investigate the metabolic changes occurring in honeybees after experimental infection with N. ceranae. Hemolymph samples were collected at two time points, 2 and 10 day post-infection, from both uninfected and infected bees. LC-MS was used to analyze individual hemolymph samples, while NMR spectra were obtained from pooled samples. The statistical analysis revealed that the age of bees had the most significant effect on the metabolite profiles. The study identified 15 biomarkers that could distinguish between infected and uninfected bees, as well as the combined effect of age and infection. These biomarkers included carbohydrates (α/β glucose, α/β fructose, and hexosamine), amino acids (histidine and proline), dipeptides (Glu-Thr, Cys-Cys, and γ-Glu-Leu/Ile), metabolites involved in lipid metabolism (choline, glycerophosphocholine, and O-phosphorylethanolamine), and a polyamine compound (spermidine) (Fig. 2). Overall, the untargeted metabolomics-based approach used in this study provides valuable insights into the pathophysiological mechanisms underlying honeybee infection by N. ceranae.
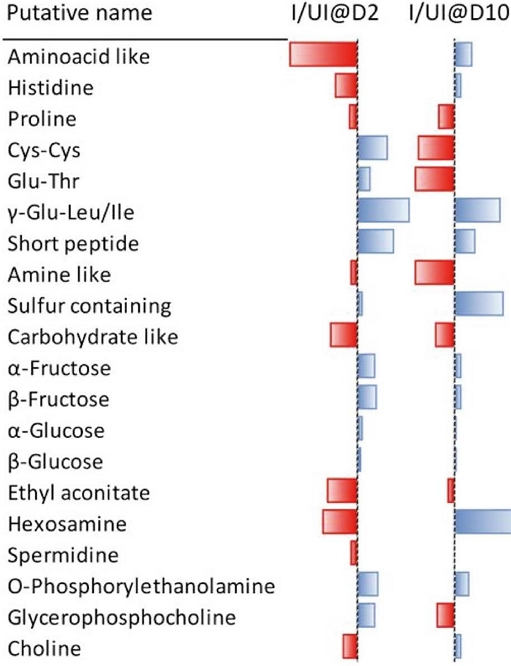
Fifteen biomarkers were successfully detected and categorized as potential biomarkers that indicate infection alone or the combined effect of age and infection. The identified biomarkers include various carbohydrates (α/β glucose, α/β fructose, and hexosamine), amino acids (histidine and proline), dipeptides (Glu-Thr, Cys-Cys, and γ-Glu-Leu/Ile), metabolites related to lipid metabolism (choline, lycerophosphocholine, and O-phosphorylethanolamine), as well as a polyamine compound (spermidine) (Jousse et al., 2020).
CONCLUSION
Metabolomics has proven to be effectively applied in various honeybee research areas, providing new insights into biology aspects such as behavior, infection, temperature stress responses, and bacteria-insect symbiosis. We are confident that this list of subjects will continue to expand in the near future. In general, it is evident that utilizing a metabolomics approach enables the identification of correlations between a phenotypic state and the underlying cellular metabolism, which older, more specific methods are unable to measure. This unique combination of comprehensive, global analysis and precise quantitative assessment presents an appealing instrument for future honeybee studies.
Acknowledgments
This work was supported by a grant from the National Institute of Agricultural Sciences, Rural Development Administration, Republic of Korea (Project No.: RS-2023-00232749).
References
-
Alaux, C., S. Sinha, L. Hasadsri, G. J. Hunt, E. Guzman-Novoa, G. DeGrandi-Hoffman, J. L. Uribe-Rubio, B. R. Southey, S. Rodriguez-Zas and G. E. Robinson. 2009. Honey bee aggression supports a link between gene regulation and behavioral evolution. Proc. Natl. Acad. Sci. U. S. A. 106(36): 15400-15405.
[https://doi.org/10.1073/pnas.0907043106]

-
Aliferis, K. A., T. Copley and S. Jabaji. 2012. Gas chromatography-mass spectrometry metabolite profiling of worker honey bee (Apis mellifera L.) hemolymph for the study of Nosema ceranae infection. J. Insect Physiol. 58(10): 1349-1359.
[https://doi.org/10.1016/j.jinsphys.2012.07.010]

-
Aliouane, Y., A. K. El Hassani, V. Gary, C. Armengaud, M. Lambin and M. Gauthier. 2009. Subchronic exposure of honeybees to sublethal doses of pesticides: effects on behavior. Environ. Toxicol. Chem. 28(1): 113-122.
[https://doi.org/10.1897/08-110.1]

-
Alonso, A., S. Marsal and A. Julia. 2015. Analytical methods in untargeted metabolomics: state of the art in 2015. Front. Bioeng. Biotechnol. 3: 23.
[https://doi.org/10.3389/fbioe.2015.00023]

-
Ardalani, H., N. H. Vidkjaer, P. Kryger, O. Fiehn and I. S. Fomsgaard. 2021. Metabolomics unveils the influence of dietary phytochemicals on residual pesticide concentrations in honey bees. Environ. Int. 152: 106503.
[https://doi.org/10.1016/j.envint.2021.106503]

-
Blacquiere, T., G. Smagghe, C. A. van Gestel and V. Mommaerts. 2012. Neonicotinoids in bees: a review on concentrations, side-effects and risk assessment. Ecotoxicology 21(4): 973-992.
[https://doi.org/10.1007/s10646-012-0863-x]

-
Blatt, J. and F. Roces. 2001. Haemolymph sugar levels in foraging honeybees (Apis mellifera carnica): dependence on metabolic rate and in vivo measurement of maximal rates of trehalose synthesis. J. Exp. Biol. 204(Pt 15): 2709-2716.
[https://doi.org/10.1242/jeb.204.15.2709]

-
Bonkowski, M. S. and D. A. Sinclair. 2016. Slowing ageing by design: the rise of NAD+ and sirtuin-activating compounds. Nat. Rev. Mol. Cell Biol. 17(11): 679-690.
[https://doi.org/10.1038/nrm.2016.93]

-
Brandt, A., A. Gorenflo, R. Siede, M. Meixner and R. Buchler. 2016. The neonicotinoids thiacloprid, imidacloprid, and clothianidin affect the immunocompetence of honey bees (Apis mellifera L.). J. Insect Physiol. 86: 40-47.
[https://doi.org/10.1016/j.jinsphys.2016.01.001]

-
Brodschneider, R. and K. Crailsheim. 2010. Nutrition and health in honey bees. Apidologie 41(3): 278-294.
[https://doi.org/10.1051/apido/2010012]

-
Brown, L. A., M. Ihara, S. D. Buckingham, K. Matsuda and D. B. Sattelle. 2006. Neonicotinoid insecticides display partial and super agonist actions on native insect nicotinic acetylcholine receptors. J. Neurochem. 99(2): 608-615.
[https://doi.org/10.1111/j.1471-4159.2006.04084.x]

-
Chakrabarti, P., J. T. Morre, H. M. Lucas, C. S. Maier and R. R. Sagili. 2019. The omics approach to bee nutritional landscape. Metabolomics 15(10): 127.
[https://doi.org/10.1007/s11306-019-1590-6]

-
Chang, H., G. Ding, G. Jia, M. Feng and J. Huang. 2022. Hemolymph Metabolism Analysis of Honey Bee (Apis mellifera L.) Response to Different Bee Pollens. Insects 14(1): 37.
[https://doi.org/10.3390/insects14010037]

-
Chen, H., K. Wang, W. Ji, H. Xu, Y. Liu, S. Wang, Z. Wang, F. Gao, Z. Lin and T. Ji. 2021. Metabolomic analysis of honey bees (Apis mellifera) response to carbendazim based on UPLC-MS. Pestic. Biochem. Physiol. 179: 104975.
[https://doi.org/10.1016/j.pestbp.2021.104975]

-
Codling, G., Y. Al Naggar, J. P. Giesy and A. J. Robertson. 2016. Concentrations of neonicotinoid insecticides in honey, pollen and honey bees (Apis mellifera L.) in central Saskatchewan, Canada. Chemosphere 144: 2321-2328.
[https://doi.org/10.1016/j.chemosphere.2015.10.135]

-
Desneux, N., A. Decourtye and J. M. Delpuech. 2007. The sublethal effects of pesticides on beneficial arthropods. Annu. Rev. Entomol. 52: 81-106.
[https://doi.org/10.1146/annurev.ento.52.110405.091440]

-
du Rand, E. E., H. Human, S. Smit, M. Beukes, Z. Apostolides, S. W. Nicolson and C. W. Pirk. 2017. Proteomic and metabolomic analysis reveals rapid and extensive nicotine detoxification ability in honeybee larvae. Insect Biochem. Mol. Biol. 82: 41-51.
[https://doi.org/10.1016/j.ibmb.2017.01.011]

-
Dussaubat, C., J. L. Brunet, M. Higes, J. K. Colbourne, J. Lopez, J. H. Choi, R. Martin-Hernandez, C. Botias, M. Cousin, C. McDonnell, M. Bonnet, L. P. Belzunces, R. F. Moritz, Y. Le Conte and C. Alaux. 2012. Gut pathology and responses to the microsporidium Nosema ceranae in the honey bee Apis mellifera. PLoS One 7(5): e37017.
[https://doi.org/10.1371/journal.pone.0037017]

-
Fischer, J., T. Muller, A. K. Spatz, U. Greggers, B. Grunewald and R. Menzel. 2014. Neonicotinoids interfere with specific components of navigation in honeybees. PLoS One 9(3): e91364.
[https://doi.org/10.1371/journal.pone.0091364]

-
Gao, J., Y. Yang, S. Ma, F. Liu, Q. Wang, X. Wang, Y. Wu, L. Zhang, Y. Liu, Q. Diao and P. Dai. 2022. Combined transcriptome and metabolite profiling analyses provide insights into the chronic toxicity of carbaryl and acetamiprid to Apis mellifera larvae. Sci. Rep. 12(1): 16898.
[https://doi.org/10.1038/s41598-022-21403-0]

-
Genersch, E. 2010. Honey bee pathology: current threats to honey bees and beekeeping. Appl. Microbiol. Biotechnol. 87(1): 87-97.
[https://doi.org/10.1007/s00253-010-2573-8]

-
Goulson, D., E. Nicholls, C. Botias and E. L. Rotheray. 2015. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 347(6229): 1255957.
[https://doi.org/10.1126/science.1255957]

-
Gray, A., R. Brodschneider, N. Adjlane, A. Ballis, V. Brusbardis, J.-D. Charrière, R. Chlebo, M. F. Coffey, B. Cornelissen, C. Amaro da Costa, T. Csáki, B. Dahle, J. Danihlík, M. M. Dražić, G. Evans, M. Fedoriak, I. Forsythe, D. de Graaf, A. Gregorc, J. Johannesen, L. Kauko, P. Kristiansen, M. Martikkala, R. Martín-Hernández, C. A. Medina-Flores, F. Mutinelli, S. Patalano, P. Petrov, A. Raudmets, V. A. Ryzhikov, N. Simon-Delso, J. Stevanovic, G. Topolska, A. Uzunov, F. Vejsnaes, A. Williams, M. Zammit-Mangion and V. Soroker. 2019. Loss rates of honey bee colonies during winter 2017/18 in 36 countries participating in the COLOSS survey, including effects of forage sources. J. Apic. Res. 58(4): 479-485.
[https://doi.org/10.1080/00218839.2019.1615661]

-
Guez, D. 2013. A common pesticide decreases foraging success and survival in honey bees: questioning the ecological relevance. Front. Physiol. 4: 37.
[https://doi.org/10.3389/fphys.2013.00037]

-
Hardie, D. G. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8(10): 774-785.
[https://doi.org/10.1038/nrm2249]

-
Henry, M., M. Beguin, F. Requier, O. Rollin, J. F. Odoux, P. Aupinel, J. Aptel, S. Tchamitchian and A. Decourtye. 2012. A common pesticide decreases foraging success and survival in honey bees. Science 336(6079): 348-350.
[https://doi.org/10.1126/science.1215039]

-
Higes, M., P. Garcia-Palencia, R. Martin-Hernandez and A. Meana. 2007. Experimental infection of Apis mellifera honeybees with Nosema ceranae (Microsporidia). J. Invertebr. Pathol. 94(3): 211-217.
[https://doi.org/10.1016/j.jip.2006.11.001]

-
Higes, M., R. Martin-Hernandez, C. Botias, E. G. Bailon, A. V. Gonzalez-Porto, L. Barrios, M. J. Del Nozal, J. L. Bernal, J. J. Jimenez, P. G. Palencia and A. Meana. 2008. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 10(10): 2659-2669.
[https://doi.org/10.1111/j.1462-2920.2008.01687.x]

-
Highfield, A. C., A. El Nagar, L. C. Mackinder, L. M. Noel, M. J. Hall, S. J. Martin and D. C. Schroeder. 2009. Deformed wing virus implicated in overwintering honeybee colony losses. Appl. Environ. Microbiol. 75(22): 7212-7220.
[https://doi.org/10.1128/AEM.02227-09]

-
Hrassnigg, N., B. Leonhard and K. Crailsheim. 2003. Free amino acids in the haemolymph of honey bee queens (Apis mellifera L.). Amino Acids 24(1-2): 205-212.
[https://doi.org/10.1007/s00726-002-0311-y]

-
Jousse, C., C. Dalle, A. Abila, M. Traikia, M. Diogon, B. Lyan, H. El Alaoui, C. Vidau and F. Delbac. 2020. A combined LC-MS and NMR approach to reveal metabolic changes in the hemolymph of honeybees infected by the gut parasite Nosema ceranae. J. Invertebr. Pathol. 176: 107478.
[https://doi.org/10.1016/j.jip.2020.107478]

-
Klein, A. M., B. E. Vaissiere, J. H. Cane, I. Steffan-Dewenter, S. A. Cunningham, C. Kremen and T. Tscharntke. 2007. Importance of pollinators in changing landscapes for world crops. Proc. Biol. Sci. 274(1608): 303-313.
[https://doi.org/10.1098/rspb.2006.3721]

-
Kostal, V., P. Simek, H. Zahradnickova, J. Cimlova and T. Stetina. 2012. Conversion of the chill susceptible fruit fly larva (Drosophila melanogaster) to a freeze tolerant organism. Proc. Natl. Acad. Sci. U. S. A. 109(9): 3270-3274.
[https://doi.org/10.1073/pnas.1119986109]

-
Kostal, V., H. Zahradnickova, P. Simek and J. Zeleny. 2007. Multiple component system of sugars and polyols in the overwintering spruce bark beetle, Ips typographus. J. Insect Physiol. 53(6): 580-586.
[https://doi.org/10.1016/j.jinsphys.2007.02.009]

-
Kunc, M., P. Dobes, R. Ward, S. Lee, R. Cegan, S. Dostalkova, K. Holusova, J. Hurychova, S. Elias, E. Pindakova, E. Cukanova, J. Prodelalova, M. Petrivalsky, J. Danihlik, J. Havlik, R. Hobza, K. Kavanagh and P. Hyrsl. 2023. Omics-based analysis of honey bee (Apis mellifera) response to Varroa sp. parasitisation and associated factors reveals changes impairing winter bee generation. Insect Biochem. Mol. Biol. 152: 103877.
[https://doi.org/10.1016/j.ibmb.2022.103877]

-
Lankadurai, B. P., E. G. Nagato and M. J. Simpson. 2013. Environmental metabolomics: an emerging approach to study organism responses to environmental stressors. Environ. Rev. 21(3): 180-205.
[https://doi.org/10.1139/er-2013-0011]

-
Le Conte, Y. and M. Navajas. 2008. Climate change: impact on honey bee populations and diseases. Rev. Sci. Tech. 27(2): 485-497, 499-510. https://www.ncbi.nlm.nih.gov/pubmed/18819674, .
[https://doi.org/10.20506/rst.27.2.1819]

-
Lee, S., F. Kalcic, I. F. Duarte, D. Titera, M. Kamler, P. Mrna, P. Hyrsl, J. Danihlik, P. Dobes, M. Kunc, A. Pudlo and J. Havlik 2022. 1H NMR Profiling of Honey Bee Bodies Revealed Metabolic Differences between Summer and Winter Bees. Insects 13(2): 193.
[https://doi.org/10.3390/insects13020193]

-
Li, Z., M. Hou, Y. Qiu, B. Zhao, H. Nie and S. Su. 2020. Changes in Antioxidant Enzymes Activity and Metabolomic Profiles in the Guts of Honey Bee (Apis mellifera) Larvae Infected with Ascosphaera apis. Insects 11(7): 419.
[https://doi.org/10.3390/insects11070419]

-
Liu, X. and J. W. Locasale. 2017. Metabolomics: A Primer. Trends Biochem. Sci. 42(4): 274-284.
[https://doi.org/10.1016/j.tibs.2017.01.004]

-
Madeira, M. S., C. Cardoso, P. A. Lopes, D. Coelho, C. Afonso, N. M. Bandarra and J. A. M. Prates. 2017. Microalgae as feed ingredients for livestock production and meat quality: A review. Livest. Sci. 205: 111-121.
[https://doi.org/10.1016/j.livsci.2017.09.020]

-
Mayack, C. and D. Naug. 2010. Parasitic infection leads to decline in hemolymph sugar levels in honeybee foragers. J. Insect Physiol. 56(11): 1572-1575.
[https://doi.org/10.1016/j.jinsphys.2010.05.016]

-
Morfin, N., T. A. Fillier, T. H. Pham, P. H. Goodwin, R. H. Thomas and E. Guzman-Novoa. 2022. First insights into the honey bee (Apis mellifera) brain lipidome and its neonicotinoid-induced alterations associated with reduced self-grooming behavior. J. Adv. Res. 37: 75-89.
[https://doi.org/10.1016/j.jare.2021.08.007]

-
Oliveira, R. A., T. C. Roat, S. M. Carvalho and O. Malaspina. 2014. Side-effects of thiamethoxam on the brain andmidgut of the africanized honeybee Apis mellifera (Hymenopptera: Apidae). Environ. Toxicol. 29(10): 1122-1133.
[https://doi.org/10.1002/tox.21842]

-
Overgaard, J., A. Malmendal, J. G. Sorensen, J. G. Bundy, V. Loeschcke, N. C. Nielsen and M. Holmstrup. 2007. Metabolomic profiling of rapid cold hardening and cold shock in Drosophila melanogaster. J. Insect Physiol. 53(12): 1218-1232.
[https://doi.org/10.1016/j.jinsphys.2007.06.012]

-
Page, R. E., Jr. and C. Y. Peng. 2001. Aging and development in social insects with emphasis on the honey bee, Apis mellifera L. Exp. Gerontol. 36(4-6): 695-711.
[https://doi.org/10.1016/s0531-5565(00)00236-9]

-
Paray, B. A., I. Kumari, Y. A. Hajam, B. Sharma, R. Kumar, M. F. Albeshr, M. A. Farah and J. M. Khan. 2021. Honeybee nutrition and pollen substitutes: A review. Saudi J. Biol. Sci. 28(1): 1167-1176.
[https://doi.org/10.1016/j.sjbs.2020.11.053]

-
Potts, S. G., V. Imperatriz-Fonseca, H. T. Ngo, M. A. Aizen, J. C. Biesmeijer, T. D. Breeze, L. V. Dicks, L. A. Garibaldi, R. Hill, J. Settele and A. J. Vanbergen. 2016. Safeguarding pollinators and their values to human well-being. Nature 540(7632): 220-229.
[https://doi.org/10.1038/nature20588]

-
Rajan, A. and N. Perrimon. 2013. Of flies and men: insights on organismal metabolism from fruit flies. BMC Biol. 11: 38.
[https://doi.org/10.1186/1741-7007-11-38]

-
Rand, E. E. D., S. Smit, M. Beukes, Z. Apostolides, C. W. W. Pirk and S. W. Nicolson. 2015. Detoxification mechanisms of honey bees (Apis mellifera) resulting in tolerance of dietary nicotine. Sci. Rep. 5: 11779.
[https://doi.org/10.1038/srep11779]

-
Requier, F., J.-F. Odoux, M. Henry, V. Bretagnolle and R. Rader. 2017. The carry-over effects of pollen shortage decrease the survival of honeybee colonies in farmlands. J. Appl. Ecol. 54(4): 1161-1170.
[https://doi.org/10.1111/1365-2664.12836]

-
Ricigliano, V. A., K. B. Cank, D. A. Todd, S. L. Knowles and N. H. Oberlies. 2022. Metabolomics-Guided Comparison of Pollen and Microalgae-Based Artificial Diets in Honey Bees. J. Agric. Food Chem. 70(31): 9790-9801.
[https://doi.org/10.1021/acs.jafc.2c02583]

-
Sanchez-Hernandez, L., D. Hernandez-Dominguez, M. T. Martin, M. J. Nozal, M. Higes and J. L. Bernal Yague. 2016. Residues of neonicotinoids and their metabolites in honey and pollen from sunflower and maize seed dressing crops. J. Chromatogr. A 1428: 220-227.
[https://doi.org/10.1016/j.chroma.2015.10.066]

-
Schafer, R. B., P. C. von der Ohe, J. Rasmussen, B. J. Kefford, M. A. Beketov, R. Schulz and M. Liess. 2012. Thresholds for the effects of pesticides on invertebrate communities and leaf breakdown in stream ecosystems. Environ. Sci. Technol. 46(9): 5134-5142.
[https://doi.org/10.1021/es2039882]

-
Shi, T., S. Burton, Y. Wang, S. Xu, W. Zhang and L. Yu. 2018. Metabolomic analysis of honey bee, Apis mellifera L. response to thiacloprid. Pestic. Biochem. Physiol. 152: 17-23.
[https://doi.org/10.1016/j.pestbp.2018.08.003]

-
Shi, X., J. Shi, L. Yu and X. Wu. 2023. Metabolic profiling of Apis mellifera larvae treated with sublethal acetamiprid doses. Ecotoxicol. Environ. Saf. 254: 114716.
[https://doi.org/10.1016/j.ecoenv.2023.114716]

-
Simon-Delso, N., V. Amaral-Rogers, L. P. Belzunces, J. M. Bonmatin, M. Chagnon, C. Downs, L. Furlan, D. W. Gibbons, C. Giorio, V. Girolami, D. Goulson, D. P. Kreutzweiser, C. H. Krupke, M. Liess, E. Long, M. McField, P. Mineau, E. A. D. Mitchell, C. A. Morrissey, D. A. Noome, L. Pisa, J. Settele, J. D. Stark, A. Tapparo, H. Van Dyck, J. Van Praagh, J. P. Van der Sluijs, P. R. Whitehorn and M. Wiemers. 2015. Systemic insecticides (neonicotinoids and fipronil): trends, uses, mode of action and metabolites. Environ. Sci. Pollut. Res. 22(1): 5-34.
[https://doi.org/10.1007/s11356-014-3470-y]

-
Simon-Delso, N., G. San Martin, E. Bruneau, L. A. Minsart, C. Mouret and L. Hautier. 2014. Honeybee colony disorder in crop areas: the role of pesticides and viruses. PLoS One 9(7): e103073.
[https://doi.org/10.1371/journal.pone.0103073]

-
Steinhauer, N. A., K. Rennich, M. E. Wilson, D. M. Caron, E. J. Lengerich, J. S. Pettis, R. Rose, J. A. Skinner, D. R. Tarpy, J. T. Wilkes and D. vanEngelsdorp. 2015. A national survey of managed honey bee 2012-2013 annual colony losses in the USA: results from the Bee Informed Partnership. J. Apic. Res. 53(1): 1-18.
[https://doi.org/10.3896/ibra.1.53.1.01]

-
Tosi, S., C. Costa, U. Vesco, G. Quaglia and G. Guido. 2018. A 3-year survey of Italian honey bee-collected pollen reveals widespread contamination by agricultural pesticides. Sci. Total Environ. 615: 208-218.
[https://doi.org/10.1016/j.scitotenv.2017.09.226]

-
van Dooremalen, C., L. Gerritsen, B. Cornelissen, J. J. van der Steen, F. van Langevelde and T. Blacquiere. 2012. Winter survival of individual honey bees and honey bee colonies depends on level of Varroa destructor infestation. PLoS One 7(4): e36285.
[https://doi.org/10.1371/journal.pone.0036285]

-
vanEngelsdorp, D., D. Caron, J. Hayes, R. Underwood, M. Henson, K. Rennich, A. Spleen, M. Andree, R. Snyder, K. Lee, K. Roccasecca, M. Wilson, J. Wilkes, E. Lengerich and J. Pettis. 2015. A national survey of managed honey bee 2010-11 winter colony losses in the USA: results from the Bee Informed Partnership. J. Apic. Res. 51(1): 115-124.
[https://doi.org/10.3896/ibra.1.51.1.14]

-
vanEngelsdorp, D., J. D. Evans, C. Saegerman, C. Mullin, E. Haubruge, B. K. Nguyen, M. Frazier, J. Frazier, D. Cox-Foster, Y. Chen, R. Underwood, D. R. Tarpy and J. S. Pettis. 2009. Colony collapse disorder: a descriptive study. PLoS One 4(8): e6481.
[https://doi.org/10.1371/journal.pone.0006481]

-
vanEngelsdorp, D. and M. D. Meixner. 2010. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J. Invertebr. Pathol. 103(Suppl 1): S80-95.
[https://doi.org/10.1016/j.jip.2009.06.011]

-
Vidau, C., M. Diogon, J. Aufauvre, R. Fontbonne, B. Vigues, J. L. Brunet, C. Texier, D. G. Biron, N. Blot, H. El Alaoui, L. P. Belzunces and F. Delbac. 2011. Exposure to sublethal doses of fipronil and thiacloprid highly increases mortality of honeybees previously infected by Nosema ceranae. PLoS One 6(6): e21550.
[https://doi.org/10.1371/journal.pone.0021550]

-
Wang, B., C. Habermehl and L. Jiang 2022. Metabolomic analysis of honey bee (Apis mellifera L.) response to glyphosate exposure. Mol. Omics 18(7): 635-642.
[https://doi.org/10.1039/d2mo00046f]

-
Watanabe, M., T. Kikawada, N. Minagawa, F. Yukuhiro and T. Okuda. 2002. Mechanism allowing an insect to survive complete dehydration and extreme temperatures. J. Exp. Biol. 205(Pt 18): 2799-2802.
[https://doi.org/10.1242/jeb.205.18.2799]

-
Williams, C. M., M. D. McCue, N. E. Sunny, A. Szejner-Sigal, T. J. Morgan, D. B. Allison and D. A. Hahn. 2016. Cold adaptation increases rates of nutrient flow and metabolic plasticity during cold exposure in Drosophila melanogaster. Proc. Biol. Sci. 283(1838): 20161317.
[https://doi.org/10.1098/rspb.2016.1317]

-
Wishart, D. S., T. Jewison, A. C. Guo, M. Wilson, C. Knox, Y. Liu, Y. Djoumbou, R. Mandal, F. Aziat, E. Dong, S. Bouatra, I. Sinelnikov, D. Arndt, J. Xia, P. Liu, F. Yallou, T. Bjorndahl, R. Perez-Pineiro, R. Eisner, F. Allen, V. Neveu, R. Greiner and A. Scalbert. 2013. HMDB 3.0 - The Human Metabolome Database in 2013. Nucleic Acids Res. 41(Database issue): D801-807.
[https://doi.org/10.1093/nar/gks1065]

-
Wishart, D. S., D. Tzur, C. Knox, R. Eisner, A. C. Guo, N. Young, D. Cheng, K. Jewell, D. Arndt, S. Sawhney, C. Fung, L. Nikolai, M. Lewis, M.-A. Coutouly, I. Forsythe, P. Tang, S. Shrivastava, K. Jeroncic, P. Stothard, G. Amegbey, D. Block, D. D. Hau, J. Wagner, J. Miniaci, M. Clements, M. Gebremedhin, N. Guo, Y. Zhang, G. E. Duggan, G. D. Macinnis, A. M. Weljie, R. Dowlatabadi, F. Bamforth, D. Clive, R. Greiner, L. Li, T. Marrie, B. D. Sykes, H. J. Vogel and L. Querengesser. 2007. HMDB: the Human Metabolome Database. Nucleic Acids Res. 35(Database issue): D521-526.
[https://doi.org/10.1093/nar/gkl923]

-
Wu, J. L., C. X. Zhou, P. J. Wu, J. Xu, Y. Q. Guo, F. Xue, A. Getachew and S. F. Xu. 2017. Brain metabolomic profiling of eastern honey bee (Apis cerana) infested with the mite Varroa destructor. PLoS One 12(4): e0175573.
[https://doi.org/10.1371/journal.pone.0175573]

-
Zhang, Z., X. Mu, Y. Shi and H. Zheng. 2022. Distinct Roles of Honeybee Gut Bacteria on Host Metabolism and Neurological Processes. Microbiol. Spectr. 10(2): e0243821.
[https://doi.org/10.1128/spectrum.02438-21]

-
Zheng, H., J. E. Powell, M. I. Steele, C. Dietrich and N. A. Moran. 2017. Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. U. S. A. 114(18): 4775-4780.
[https://doi.org/10.1073/pnas.1701819114]

