Regulation of mRNA Expression in Head and Gut of Apis mellifera after Nosema ceranae Infection
Abstract
Honeybees (Apis mellifera) have been threatened by diseases and their numbers have been noticeably declining. Nosemosis, a disease caused by Nosema ceranae, is now a leading threat to honeybees. N. ceranae is a microsporidian that forms a spore before it infects the host, which makes it especially difficult to control compared to other pathogens. Because the awareness of nosemosis risk increased within decades, N. ceranae has been studied as a pathogen. Some studies showed the up/down-regulation of honeybee mRNA after nosemosis, however, there are no definite mRNAs that can be used as markers for the disease. In this study, to better understand the honeybee’s regulation of mRNA expression after infection by N. ceranae and to determine a marker for nosemosis, 48 mRNAs from the honeybee’s metabolic pathway, immune system, hormone, or unknown mRNAs, were collected from honeybee head and gut and were quantified using real-time PCR. We selected 18 candidates with the largest expression changes and confirmed that more genes from the metabolic pathway were regulated after infection than immune-related genes. Also noteworthy is that the tendency to regulate gene expression in the head and gut differed. In conclusion, many genes, including Aminopeptidase1, GB50026, and Trypsin alpha can be reliable markers for nosemosis, as there are huge differences in the number of transcripts according to the body part.
Keywords:
Nosemosis, Nosema ceranae, Apis mellifera, Infection, MarkerINTRODUCTION
Honeybees (Apis mellifera) are not only essential in crop and fruit industries, but also critical to our ecosystem as pollinators (Papa et al., 2022). Considering their contribution as a defender of our ecosystem, their economic value was estimated as $14.6 billion in 2000 (Morse and Calderone, 2000), which has continued to increase. However, this valuable insect is now threatened by the several pathogens to which they are exposed. Recently, their colonies have disappeared more often, and the term colony collapse disorder (CCD) is appearing more frequently in papers than ever before (Vanengelsdorp et al., 2009). The root cause of CCD remains unknown however, honeybee pathogens are plausible contributors (Vanengelsdorp et al., 2017). They can be spread outside the colonies since adult bees leave their colonies to search for the source of food. However, the most plausible contributor among honeybee pathogens is Nosema (Higes et al., 2008; Ellis et al., 2010; Dainat et al., 2012).
Nosema ceranae is a kind of microsporidia, a spore- forming fungus that causes nosemosis (Fries, 2010). Once N. ceranae enters the honeybee by ingestion, it infects honeybee midgut cells (Kurze et al., 2018). Upon germination inside the host, they start to change the honeybee metabolism and proliferate until they are fully grown. The fungus takes about five days to fully grow in a honeybee gut cell, and after 20 days, the honeybee starts to die because of the disease (Milbrath et al., 2015). It is highly infectious to honeybees and now it is well known to be globally distributed (Dainat et al., 2012). Also, they were recently found in larvae of honeybees, meaning that they can infect any stage of honeybees (Eiri et al., 2015). Although the guts are the place where nosemosis take place, the brain serves as the center of the nervous system in the honeybees. The gene expression changes in the brain are important for behavioral plasticity and the decision to change in response to external stimuli (Robinson et al., 2008).
Since the initial discovery of N. ceranae, researcher have found some genetic markers in honeybees that can be used to determine if nosemosis is within the colony (Antúnez et al., 2009). However, the information is only limited to the genes of immunoproteins, which is not sufficient to fully understand the complete mechanism of disease progression in honeybees. To do so, the transcriptomic approach in gene expression regardless of the category of the genes caused by nosemosis and their effect on honeybees must be fully studied.
To understand gene expression regulation in honeybees with nosemosis, qPCR was done with 48 mRNAs (Table 1), and the 18 most altered genes were identified (Table 2). Considering different mechanisms of gene expression may be present in different body locations, samples were obtained from the head and gut, individually, and results were statically analysed to compare differences after infection.
METHODS AND MATERIALS
1. Spore preparation
N. ceranae-infected honeybees were obtained from the Rural Development Administration in Korea (RDA, Korea). Midgut tissues of infected bees were homogenized in 200 μL of distilled water to obtain N. ceranae spores for inoculations. N. ceranae was purified from the suspension using discontinuous 25%, 50%, 75%, and 90% Percoll (GE healthcare, USA) (Kim et al., 2017). The spore concentration was determined using a hemocytometer; spores were subsequently mixed with 50% sucrose solution to achieve 20,000 spores/ μL.
2. A. mellifera handling
Nosema spores were injected into bees that had recently emerged, instead of bees in their late stage, because new emerging bees do not require the use of carbon dioxide and more accurate results can be achieved (Milbrath et al., 2015). After emergence, 15 to 20 honeybees were divided in to two groups. Honeybees from one group were starved for two hours and then fed a 3 μL 50% sucrose solution with 60,000 Nosema spores (Malone et al., 1995). Honeybees from another group were treated in the same process using a 50% sucrose solution without spores. Bees were grown in a cage separately with dark at 28℃. After a 14-day growth period, all bees were stored at -70℃ until use.
3. RNA isolation and cDNA synthesis
The midgut and head of the honeybees from each group were separated using a sterilized blade. The parts were disrupted using a mortar and pestle in 3 mL of RIPA buffer. DNAs were eliminated using DNase (Thermo, USA) treatment, and RNAs from the obtained suspension were extracted using an RNA mini kit (Ambion, USA). Total RNAs from each group were adjusted to the same concentration and cDNA was synthesized using reverse transcriptase (Thermo, USA).
4. Quantification of transcript using real-time PCR
Using synthesized cDNA as a template with qPCR master mix (Cellsafe, USA) and primers (Table 3), realtime PCR was performed. For initial denaturation, we heated the suspension at 95℃ for five minutes. For the PCR cycle, the following steps were repeated 28 times: 95℃ for 20 seconds, 49-58℃ for 20 seconds, and 72℃ for 20 seconds. As a final elongation step, 72℃ was maintained for five minutes, followed by 4℃ maintenance. Real-time PCR was carried out using CFX Connect (Bio-Rad, USA). Once all genes were screened, we identified 18 genes with large differences and repeated real-time PCR on those candidates six times.
5. Normalization of real-time data and statistical analysis
Results from real-time PCR were expressed as a Cq value, which denotes the number of cycles required to generate a signal greater than the threshold. All assay results were normalized to accurately compare with other genes (Vandesompele et al., 2002). For reference, the transcript level of actin was measured and transcript normalization used the following equation.
6. Statistical analysis
Results are presented as the mean±mean standard deviation (S.D). To determine the reproducibility of the measurements, each qRT-PCR assay was carried out in triplicate. Statistical analysis was performed using Microsoft Excel 2021 (Microsoft, USA) and Statistical Package for the Social Sciences (SPSS) 28.0 (IBM Corp, USA). All graphs except microscopic images were analyzed by Microsoft Excel 2021. Statistical significance was defined at a p-value less than 0.05.
RESULTS
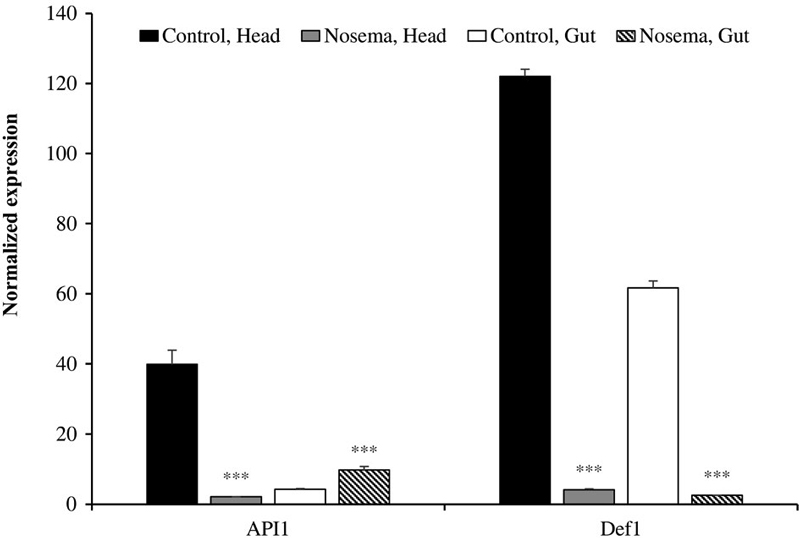
1. Quantification of apidaecin and defensin
Since apidaecin (API1) and defensin (Def1) were already identified as genes upregulated upon nosemosis infection (Casteels et al., 1989; Klaudiny et al., 2005), they were measured first. Expressions of both genes from the head were dramatically repressed after infection (Fig. 1). Normalized values for API1 and Def1 from the head were 39.9±4.1 and 122±2 but changed to 2.1±0.1 and 4.1±0.3, respectively. However, the tendency to change was not the same in gut cells. While expression of apidaecin was slightly increased from 4.2±0.2 to 9.7±1, that of defensin showed a significant decrease from 61.7±2 to 2.5±0.1 after infection. N. ceranae affected the expression of defensin in the honeybee immune system.
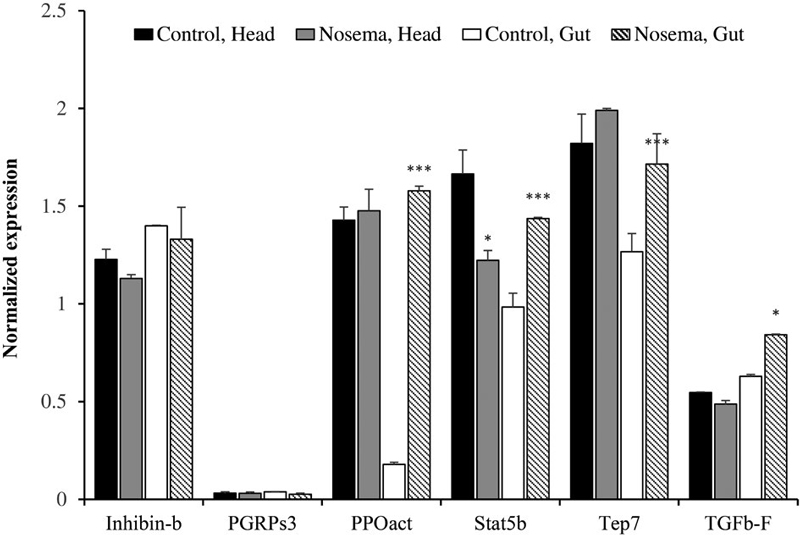
2. Quantification of immune-related genes
Aside from apidaecin and defensin, we sorted for immune-related candidate genes which can be used as genetic markers for nosemosis (Fig. 2). Those genes were inhibin beta (Inhibin-b), peptidoglycan recognition protein (PGRPs3), PPOact, signal transducer and activators of transcription (Stat5b), CD109 (Tep7), and transforming growth factor beta (TGFb-F). Inhibin-b and PGRPs3 did not show significant changes in their expression. There were no large changes in the amount of PPOact from the head, however, midgut cells showed dramatic differences from 0.18±0.01 to 1.58±0.25. Tep7 and TGFb-F transcripts from the head also not changed, while the same transcript from gut cells increased. In the case of Stat5b, its transcription level from both body parts changed. The amount of Stat5b from the head changed from 1.66±0.12 to 1.22±0.05, and from 0.98±0.07 to 1.44±0.01 in the gut. Transcription level of most immune-related genes was induced, implying that the honeybee immune system was activated after nosemosis infection.
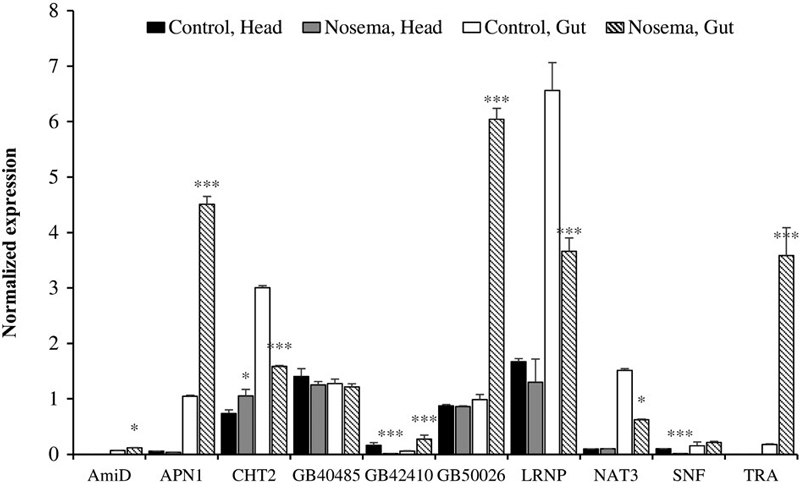
3. Quantification of non-immune genes
Parasitic characteristics of N. ceranae could change the gene expression of the metabolic pathway of honeybee (Mayack and Naug, 2009). As a result, we found nonimmune genes with significant changes in transcript level (Fig. 3). Those genes were N-acetylmuramoyl-L-alanine amidase (AmiD), aminopeptidase1 (APN1), chymotrypsin2 (CHT2), GB40485, GB42410, GB50026, leucinerich repeat neuronal protein (LRNP), natterin3 (NAT3), short neuropeptide F-like (SNF), and trypsin alpha (TRA). The transcript expression rate of AmiD and GB40485 changed only slightly compared to other non-immune genes. Among those candidate genes, two of them oppositely changed their expression. The transcription level of CHT2 from the head increased while that from the gut decreased. On the other hand, the transcription level of GB42410 from the head decreased while that from the gut increased. APN1, GB50026, LRNP, NAT3, and TRA changed their transcription level only in gut cells, and except NAT3, significantly large expression changes were noted. APN1 changed from 1.05±0.02 to 4.51± 0.14, GB50026 changed from 0.99±0.09 to 6.04±0.20, LRNP changed from 6.56±0.50 to 3.66±0.24, and TRA changed from 0.18±0.01 to 3.58±0.51. SNF changed its expression only in the head, from 0.10±0.002 to 0.011±0.0008. Although both APN1 and CHT2 are related to the digestive system, they seemed to respond differently against nosemosis.
DISCUSSION
The disastrous effect, that nosemosis could bring on apiculture or even on our ecosystem has been repeatedly reported (Klee et al., 2007; Higes et al., 2010). Also, after nosemosis was blamed for being the cause of CCD, studies on Nosema spp. progressed rapidly. Many studies reported changes in gene transcription levels in honeybees after nosemosis, and some listed immune genes altered after infection (Antúnez et al., 2009; Chaimanee et al., 2012). Here, we found genes that showed altered expression whether they are related to immune function or not, and have reported results by expression location.
Using quantitative real-time PCR, we confirmed that more genes from the metabolic pathway were regulated after infection than immune-related genes. As previously described, it was undoubtedly true that apidaecin1 and defensin1 changed the most in terms of transcription numbers and rates (Casteels et al., 1989; Klaudiny et al., 2005). The transcription level of those genes was greatly changed, both in the head and gut. Since both genes showed large changes, this suggests they are the best genetic markers for nosemosis. Surprisingly, the expression of apidaecin1 was down-regulated in head and for defensin1, its expression was down-regulated in both head and gut. Considering they are well established immunoprotein, they were expected to be up-regulated. According to previous studies, the time and the number of spores fed to honeybees are important factors for the expression, and also, decreased expression of immunoproteins in nosemosis-infected honeybees are reported (Chaimanee et al., 2012). To understand honeybee immune mechanisms and the parasitic characteristics of N. ceranae, we needed more understanding about other genes related to N. ceranae infection that are not well established.
Our results showed that most immune-related genes from honeybee gut cells were increased in expression upon N. ceranae infection. Although honeybees were infected with N. ceranae for 14 days, which is right before honeybee death began, their immune system was still functioning. These results may lead us to a special mechanism, that N. ceranae uses, to survive against these immune responses. It can be debatable that those genes except apidaecin1 and defensin1, which were previously studied as induced gene expression after nosemosis, are regulated differently by the microbiome of honeybees since the microbiome of honeybees are influenced by nosemosis and not discovered in previous nosemosis studies (Huang et al., 2018). The cause of gene expressions of immunoproteins can result from changes in the microbiome, however, either way, it is undoubtedly clear that nosemosis affects guts and immune systems of honeybees.
Results presented on expression differences of nonimmune genes implied that N. ceranae alters the gut cell system to help spore survival inside honeybee gut cells. Expression of genes related to many digestive systems was decreased and expression of genes that provide nutrition to cells was increased. Considering N. ceranae is a parasite-like microsporidian, the spores may promote honeybee guts to be changed as a suitable environment for them, by inducing the provision of nutrition they need and reducing the loss of nutrition they can use (Paris et al., 2018). The observed alterations in gene expression within the heads of the bees could suggest a close relationship between the environment in which N. ceranae thrives and these specific genes. This is further supported by the fact that gene expression within the head of bee either remained stable or exhibited only minor changes.
It is important to note that the transcription rates of GB42410 and GB50026 were changed. Although their function is still unknown, we expect GB42410 is to contribute to the immune system, and GB50026 is to contribute to the metabolic pathway. Keeping up with the descriptions above, the expression of GB42410 has changed in both the head and gut of honeybees, which means GB42410 may be closely related to the immune system. On the other hand, the transcription level of GB50026 increased almost six times in the midgut of honeybees, and it is similar to the tendency of nonimmune genes above. Also, great changes in the expression of GB50026 could mean that it is an important target gene for N. ceranae. Those unknown genes should be studied further for an improved understanding of honeybee infection. Also, as an extension of these findings, changes in transcription level caused by nosemosis using honeybees from egg to their late stage will be studied, since nosemosis can occur in any stage of honeybees. It will provide us more understanding of epigenetic changes in immune systems and it may even provide a breakthrough in nosemosis by determining possible treatment stages for the complete cure. Along with epigenetic studies, the proteins mentioned above will be treated in honeybees in further studies to determine whether the addition of extra genes may help prevent or also cure N. ceranae infection.
Acknowledgments
This work was supported by the National R&D Program through the National Research Foundation of Korea (NRF) funded by the Korea government (Ministry of Science and ICT) (2021R1A6A3A13046056).
FOOTNOTES
Financial Disclosure: This research was conducted in the absence of any commercial or financial relationships that could be construed as posing potential conflicts of interest.
References
-
Antúnez, K., R. Martín-Hernández, L. Prieto, A. Meana, P. Zunino and M. Higes. 2009. Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia). Environ. Microbiol. 11(9): 2284-2290.
[https://doi.org/10.1111/j.1462-2920.2009.01953.x]

-
Casteels, P., C. Ampe, F. Jacobs, M. Vaeck and P. Tempst. 1989. Apidaecins: antibacterial peptides from honeybees. EMBO J. 8(8): 2387-2391.
[https://doi.org/10.1002/j.1460-2075.1989.tb08368.x]

-
Chaimanee, V., P. Chantawannakul, Y. Chen, J. D. Evans and J. S. Pettis. 2012. Differential expression of immune genes of adult honey bee (Apis mellifera) after inoculated by Nosema ceranae. J. Insect Physiol. 58(8): 1090-1095.
[https://doi.org/10.1016/j.jinsphys.2012.04.016]

-
Dainat, B., D. Vanengelsdorp and P. Neumann. 2012. Colony collapse disorder in Europe. Environ. Microbiol. Rep. 4(1): 123-125.
[https://doi.org/10.1111/j.1758-2229.2011.00312.x]

-
Eiri, D. M., G. Suwannapong, M. Endler and J. C. Nieh. 2015. Nosema ceranae can infect honey bee larvae and reduces subsequent adult longevity. PLoS One 10(5): e0126330.
[https://doi.org/10.1371/journal.pone.0126330]

-
Ellis, J. D., J. D. Evans and J. Pettis. 2010. Colony losses, man- aged colony population decline, and colony collapse dis- order in the United States. J. Apic. Res. 49(1): 134-136.
[https://doi.org/10.3896/IBRA.1.49.1.30]

-
Fries, I. 2010. Nosema ceranae in European honey bees (Apis mellifera). J. Invertebr. Pathol. 103: S73-S79.
[https://doi.org/10.1016/j.jip.2009.06.017]

-
Higes, M., R. Martín-Hernández and A. Meana. 2010. Nosema ceranae in Europe: an emergent type C nosemosis. Api- dologie 41(3): 375-392.
[https://doi.org/10.1051/apido/2010019]

-
Higes, M., R. Martín-Hernández, C. Botías, E. G. Bailón, A. V. González-Porto, L. Barrios, M. J. Del Nozal, J. L. Bernal, J. J. Jiménez, P. G. Palencia and A. Meana. 2008. How natural infection by Nosema ceranae causes honeybee colony collapse. Environ. Microbiol. 10(10): 2659-2669.
[https://doi.org/10.1111/j.1462-2920.2008.01687.x]

-
Huang, S. K., K. T. Ye, W. F. Huang, B. H. Ying, X. Su, L. H. Lin, J. H. Li, Y. P. Chen, J. L. Li, X. L. Bao and J. Z. Hu. 2018. Influence of feeding type and Nosema ceranae infection on the gut microbiota of Apis cerana workers. mSystems 3(6): e00112-18.
[https://doi.org/10.1128/mSystems.00177-18]

-
Kim, D. J., H. G. Yun, I. H. Kim, W. S. Gwak and S. D. Woo. 2017. Efficient method for the rapid purification of Nosema ceranae spores. Mycobiology 45(3): 204-208.
[https://doi.org/10.5941/MYCO.2017.45.3.204]

-
Klaudiny, J., Š. Albert, K. Bachanová, J. Kopernický and J. Šimúth. 2005. Two structurally different defensin genes, one of them encoding a novel defensin isoform, are expressed in honeybee Apis mellifera. Insect Biochem. Mol. Biol. 35(1): 11-22.
[https://doi.org/10.1016/j.ibmb.2004.09.007]

-
Klee, J., A. M. Besana, E. Genersch, S. Gisder, A. Nanetti, D. Q. Tam, T. X. Chinh, F. Puerta, J. M. Ruz, P. Kryger and D. Message. 2007. Widespread dispersal of the microspo- ridian Nosema ceranae, an emergent pathogen of the western honey bee, Apis mellifera. J. Invertebr. Pathol. 96(1): 1-10.
[https://doi.org/10.1016/j.jip.2007.02.014]

-
Kurze, C., Y. Le Conte, P. Kryger, O. Lewkowski, T. Müller and R. F. Moritz. 2018. Infection dynamics of Nosema ceranae in honey bee midgut and host cell apoptosis. J. Invertebr. Pathol. 154: 1-4.
[https://doi.org/10.1016/j.jip.2018.03.008]

-
Malone, L. A., H. A. Giacon and M. R. Newton. 1995. Compar- ison of the responses of some New Zealand and Aus- tralian honey bees (Apis mellifera L) to Nosema apis Z. Apidologie 26(6): 495-502.
[https://doi.org/10.1051/apido:19950606]

-
Mayack, C. and D. Naug. 2009. Energetic stress in the honey- bee Apis mellifera from Nosema ceranae infection. J. Invertebr. Pathol. 100(3): 185-188.
[https://doi.org/10.1016/j.jip.2008.12.001]

-
Milbrath, M. O., T. van Tran, W. F. Huang, L. F. Solter, D. R. Tarpy, F. Lawrence and Z. Y. Huang. 2015. Compara- tive virulence and competition between Nosema apis and Nosema ceranae in honey bees (Apis mellifera). J. Invertebr. Pathol. 125: 9-15.
[https://doi.org/10.1016/j.jip.2014.12.006]

- Morse, R. A. and N. W. Calderone. 2000. The value of honey bees as pollinators of US crops in 2000. Bee Cult. 128(3): 1-15.
-
Papa, G., R. Maier, A. Durazzo, M. Lucarini, I. K. Karabagias, M. Plutino, E. Bianchetto, R. Aromolo, G. Pignatti, A. Ambrogio and M. Pellecchia. 2022. The honey bee Apis mellifera: An insect at the interface between human and ecosystem health. Biology 11(2): 233.
[https://doi.org/10.3390/biology11020233]

-
Paris, L., H. El Alaoui, F. Delbac and M. Diogon. 2018. Effects of the gut parasite Nosema ceranae on honey bee physi- ology and behavior. Curr. Opin. Insect Sci. 26: 149-154.
[https://doi.org/10.1016/j.cois.2018.02.017]

-
Robinson, G. E., R. D. Fernald and D. F. Clayton. 2008. Genes and social behavior. Science 322(5903): 896-900.
[https://doi.org/10.1126/science.1159277]

-
Vandesompele, J., K. De Preter, F. Pattyn, B. Poppe, N. Van Roy, A. De Paepe and F. Speleman. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3(7): research0034-1.
[https://doi.org/10.1186/gb-2002-3-7-research0034]

-
Vanengelsdorp, D., J. D. Evans, C. Saegerman, C. Mullin, E. Haubruge, B. K. Nguyen, M. Frazier, J. Frazier, D. Cox-Foster, Y. Chen and R. Underwood. 2009. Colony collapse disorder: a descriptive study. PLoS One 4(8): e6481.
[https://doi.org/10.1371/journal.pone.0006481]

-
Vanengelsdorp, D., K. S. Traynor, M. Andree, E. M. Lichtenberg, Y. Chen, C. Saegerman and D. L. Cox-Foster. 2017. Colony Collapse Disorder (CCD) and bee age impact honey bee pathophysiology. PLoS One 12(7): e0179535.
[https://doi.org/10.1371/journal.pone.0179535]