Novel Bacteriocins from Geobacillus thermoleovorans and G. stearothermophilus against Paenibacillus larvae
Abstract
American Foulbrood caused by Paenibacillus larvae is the most destructive disease of honey bee. This study aimed at investigating the growth inhibition potential of bacteriocins originated from Geobacillus thermoleovorans (GT-1326) and G. stearothermophilus (GS-1368) on P. larvae based on in vitro and in silico approach. The results showed the minimum inhibition concentration values of the both bacteriocins were the same as 800 µg/mL. Moreover, the cell leakage assay demonstrated the increase of nucleic acid amount observed within the cell-free supernatant in dose-dependent manner. Furthermore, the in silico approach predicted that the bacteriocins structure models comprised of A chain and six α-helix that enables to penetrate P. larvae membrane. The physicochemical properties data confirmed coomassie blue staining results in terms of the size of bacteriocins’ molecule (approximately 32 kDa) with the instability index reached 50 and hydrophobicity under threshold value. Additionally, the interactions occurred between bacteriocins and target protein were dominated by hydrophobic bonds. The prediction of bacteriocin mechanism of action depicted that the function was close to iron-chelating agent of bacteria. By interacting with lipoteichoic acid synthase located on the P. larvae surface, the bacteriocins compete with P. larvae to gain iron in which the pivotal nutrient for the metabolic process. Based on the inhibitory ability of GT-1326 and GS-1368 on the growth of P. larvae, further studies need to be done to increase the efficiency of the bacteriocins.
Keywords:
American Foulbrood, Bacteriocin, G. thermoleovorans, G. stearothermophilus, P. larvaeINTRODUCTION
Honey bee, Apis mellifera, plays a crucial role in ecosystem (Hung et al., 2018). It has been widely known as a generalist pollinator (Hung et al., 2018) which giving benefits for agricultural plants cultivation (Aizen et al., 2008). A. mellifera has even been reported as one of pollinators on which United-States economic-agricul- tural sector is depended (Khanna et al., 2021). Moreover, managed honey bees provided honey production valued at eight billion US dollars in 2020 (Shahbandeh, 2022). However, the emergence of highly contagious disease, American Foulbrood (AFB) (Genersch, 2010), threated honey bee life.
To date, AFB is one of the most destructive diseases of honey bee caused by Paenibacillus larvae (Fünfhaus et al., 2018). It is a gram-positive, spore-forming faculta- tive anaerobic bacterium (Ash et al., 1993) which kills the bee larvae. After killing bee larvae, P. larvae sub- sequently degrades the cadaver into dry slimy mass that so called foulbrood scale consisting P. larvae spores (Fünfhaus et al., 2018). A substantial financial losses of beekeeping industry has been predicted in accor- dance with AFB case. This due to the costs of replacing infected hives, bees, and brood, the loss revenue as the reduction in honey production, hive rental income, hives sterilization and the labor costs associated with the implementation of best practices management for AFB prevention and spread controlling (Laate et al., 2020).
Controlling AFB is still challenging at several points. As the disease cannot be cured, a procedure to conduct, in case of disease, is burning the beehives of the infected colony altogether with the bees and honey (Debeljak and Vidanovi, 2023). Antibiotics use is one of options which can be chosen to control P. larvae. However, antibiotics are not allowed to use in some countries (Genersch, 2010) such as European countries (World Animal Protection, 2022) and U.S. (AccessScience Editors, 2017). Not only does the use of antibiotics cause resistance, but it also, in turn, causes an accumulation of resistant genes in bee gut (Tian et al., 2012). Therefore, the obstacles in handling this disease are still a big issue which requires serious effort to perform.
Antimicrobial peptides and proteins are potential alternatives to deal with the issue. One of the benefits offered by utilizing these antimicrobial agents is their slow or even absent bacterial resistance rate (Sarkar et al., 2021). Among many microbial resistances against conventional antibiotics (Okamoto et al., 2021), the advantage of these nature-originated peptides and protein are promising to develop.
Acknowledging the cruciality of novel antimicrobial agent, scientists have started to investigate the antimicrobial potential possessed by Geobacillus genus. It was reported that Geobacillus strain was able to inhibit and kill Aspergillus fumigatus, Botrytis cinerea, Verticillium dahliae, and gram-negative bacteria (Zebrowska et al., 2022). The purpose of this study is to investigate the growth inhibition potential of bacteriocins from Geobacillus thermoleovorans and Geobacillus stearothermophilus against P. larvae with in vitro and in silico approaches. The findings of this research are expected to contribute in providing basic information of antimicrobial agents against P. larvae as the causative agent of AFB.
MATERIALS AND METHODS
1. Bacterial strains and growth conditions
P. larvae [ATCC 9545] (ERIC I) was purchased from American Type Culture and Collection (ATCC), meanwhile, G. thermoleovorans [KACC 11374], and G. stearothermophilus [KACC 10843] were purchased from Korean Agricultural Culture Collection (KACC). P. larvae was cultured in MYPGP media which comprised of Mueller-Hinton broth, yeast extract, potassium monoacid phosphate, glucose, and sodium pyruvate at 37℃ in with shaking for 24 hours (De Graaf et al., 2013). Meanwhile, G. thermoleovorans and G. stearothermophilus were cultured in the nutrient agar media which comprised of yeast extract, peptone, NaCl, Agar, and distilled water at the temperatures of 70℃ and 55℃, respectively.
2. Gene cloning
The bacteriocins of G. thermoleovorans was labelled with GT-1326 and the G. stearothermophilus was labelled with GS-1368. GT-1326 and GS-1368 genes were amplified by PCR using two pairs of primers. The primer pair used to amplify G. thermoleovorans were forward: 5ʹ-ATCGCCATGGATAAAACGAAGCTGTATCCA-3ʹ and reverse: 5ʹ-ATCGAAGCTTTGCTGACTCGTTCGGATCCT-3ʹ which contained NcoI and HindIII restriction sites, respectively. Meanwhile, the bacteriocin gene of G. stearothermophilus was amplified using primer pair of forward: 5ʹ-ATCGCCATGGATAAAACGAAGCTGTATCCA-3ʹ and reverse: 5ʹ-ATCGCTCGAGCGAGGCTCCGGCTGGATCTT-3ʹ which comprised of NcoI and XhoI restriction sites. The amplified DNA were cloned into pET28a plasmid vector which then transformed into Escherichia coli BL21 to express the bacteriocins.
3. Bacteriocin expression and purification
E. coli which harboring pET28a with bacteriocin genes were cultured overnight in Luria Bertani (LB) broth media with kanamycin. The cultures were then adjusted to 0.8 of OD600 and treated Isopropyl b-D-1thiogalactopyranoside (IPTG) 0.4 mM for six hours before harvesting. The harvesting process was done by centrifuging the cultures at 3,000 rpm for 10 minutes, after which the pellet was stored at -70℃ overnight. The pellet of E. coli bearing the bacteriocins were then mixed with native binding buffer (50 mM NaH2PO pH 8.0, 250 mM NaCl, 10 mM Imidazole) and lysed using sonicator (Sonics & Materials, Inc., USA) in three cycles which encompassed 15 seconds on and 15 seconds off for three minutes with the amplitude of 30%. The disrupted cells were centrifuged in 4,000 rpm for ten minutes. The supernatant was transferred into the tube with 1 mL of Ni-NTA (Qiagen, Germany). The mixture was placed in shaker at 4℃ for one hour and washed (50 mM NaH2PO4 pH 8.0, 500 mM NaCl, 20 mM Imidazole) and followed with elution process (50 mM NaH2PO4 pH 8.0, 500 mM NaCl, 250 mM Imidazole). The eluted protein was stored in -80℃ until it is used.
4. Purified bacteriocin check
To ensure the purity as well as the protein size, the 10%-SDS PAGE gel was used to load the purified protein. The separation process was set at 80 volt and ten minutes for stacking gel, and 100 volt and 1.5 hours for separating gel. The loaded protein was stained with coomassie brilliant blue.
5. Minimum inhibitory concentration
An initial test conducted to measure bacteriocins activity was Minimum Inhibitory Concentration (MIC). This assay was performed based on Clinical and Laboratory Standards Institute (CLSI) document MI00-S21 besides considering several previous studies. A 96-well plate was used to conduct microdilution in which the P. larvae in MYPGP broth medium was defined in 0.01 of OD600. The P. larvae was treated GT-1326, GS-1368 and antibiotic with two-fold dilution. The P. larvae cells were poured into 96-well plate with the final volume 100 μL per well in 11 lanes. The first lane wells were defined for only cells. Meanwhile, the second to eleventh lane wells were used to treat the P. larvae cells using bacteriocins. The bacteriocin concentrations used ranged from 6.25 to 1,000 μg/mL. As the dilutions well done, the cells were incubated in 37℃ for 24 hours. By considering the MIC value gained (i.e., 800 μg/mL), the lower and higher bacteriocin concentrations were used. The MIC procedure was repeated three times. The elution buffer was also applied as control in this procedure to ensure if the buffer interfere the antimicrobial effect on P. larvae. By considering the antifungal activity of imidazole, this experiment used diluted imidazole to ensure that the concentration will not block the P. larvae growth.
6. Cell leakage assay
Cell leakage assay was conducted to ensure if cell leakage occurred in the treated P. larvae. This assay was started by stopping P. larvae cells proliferation using PBS for two hours. Using the MIC value, lower and higher bacteriocin concentrations were tested. The cells were treated using bacteriocins with the concentrations of 250, 500, and 1,000 μg/mL for an hour. Negative controls were loaded together with the same bacteriocin concentrations in PBS to ensure that the absorbance values were not interfered by the bacteriocins. The cells were then centrifuged at 7,000 rpm for ten minutes to precipitate the cell debris. The supernatant was measured at 260 nm using an ELISA reader (microplate spectrophotometer, Epoch).
7. In silico studies
In silico studies were started with protein structure modelling of bacteriocins, both GT-1326 and GS-1368. The modelling was done by submitting the bacteriocins’ amino acid sequences to I-TASSER (https://zhanggroup.org/I-TASSER/). Based on the structure model, the analysis of receptor protein was done using literature study. The physicochemical properties in terms of molecular weight, theoretical isoelectric point (PI), grand average of hydropathicity (GRAVY), as considerable as instability index of the bacteriocins were predicted using ExPASy (http://web.expasy.org/protparam/). Lipoteichoic acid of P. larvae as the receptor of bacteriocin (Rosa et al., 2017) was determined based on the bacteriocin structure prediction results. The alpha fold model was downloaded from Uniprotkb (https://www.uniprot.org/uniprotkb) with the protein code of W2E6E6. Meanwhile, the docking process was done by using HDOCK server (http://hdock.phys.hust.edu.cn/) in which the visualization was done using BIOVIA Discovery Studio 2021 Client (Dassault Systèmes, 2020).
RESULTS
1. The purification of GT-1326 and GS-1368
The purified products were separated using SDS-PAGE and stained with coomassie brilliant blue. Fig. 1 shows the bands of bacteriocins which were observed in the size approximately 32 kDa. The bacteriocins of GT-1326 and GS-1368 were well-purified.
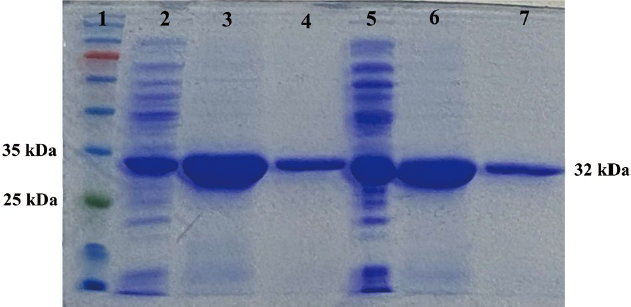
SDS-PAGE of GT-1326 and GS-1368 bacteriocins in 10% acrylamide with the loading setting of 80 v for 10 minutes (stacking gel) and 100 v for 1.5 hours (separating gel). (1) Marker; (2) Unpurified GT-1326; (3) Purified GT-1326 with first elution buffer; (4) Purified GT-1326 with second elution buffer; (5) Unpurified GS-1368; (6) GS-1368 with first elution buffer; (7) GS-1368 with second elution buffer.
2. Bacteriocins activity in inhibiting P. larvae growth by inducing membrane cell leaking
Bacteriocins activity in affecting P. larvae was initially investigated using MIC method. The results of this research are depicted in Table 1. It is obvious that the bacteriocins originated from G. thermoleovorans and G. stearothermophilus inhibited P. larvae in the minimum dosage of 800 μg/mL which were higher compared to ampicillin (25 μg/mL).
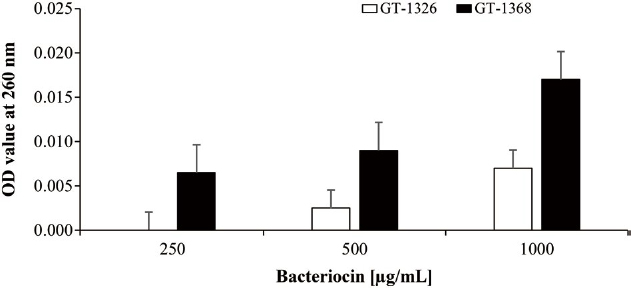
Finding an inhibition activity of the bacteriocins in MIC results, bacterial cell leakage assay was done to find basal mechanism. The measurement of nucleic acid dissolved in the cell-free supernatant (CFS) was conducted. The bar chart of Fig. 2 shows the increase of nucleic acid content in the CFS of dose-dependently treated bacteriocins against P. larvae.
3. Bacteriocin-protein interaction prediction through in silico studies
The antibacterial mechanism of bacteriocin against P. larvae is the pivotal point to clarify. Therefore, in silico docking was performed to predict the interactions between bacteriocins and protein target of P. larvae. Bacteriocins profiling was done in five steps encompassed structure modelling, physicochemical properties characterization, protein target determination, molecular docking, and protein interaction visualization.
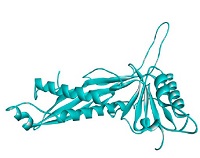
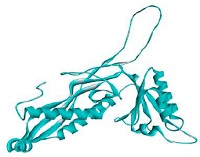
The prediction results of bacteriocins structure are served in Fig. 3. There were ten proteins found in PDB database which were used as the templates to construct bacteriocins (GT-1326 and GS-1368) structures. The resulted models are shown in Table 2. It is depicted that the tertiary structures of the both bacteriocins were A chain which encompasses six α-helix in several parts.
Meanwhile, bacteriocins characterization which was done using ProtParam computation resulted several important values of physicochemical properties i.e., molecular weight, theoretical PI, Grand Average of Hydrophaticity (GRAVY), and stability index as served in Table 2.
Based on ProtParam computation, the size of both bacteriocins, GT-1326 and GS-1368, were 31840.34 Da and 31691.22 Da respectively. These sizes were in line with the SDS-PAGE quantification results (Fig. 1) in which the size of bacteriocins were similar i.e., around 32 kDa. In addition, the hydrophobicity of bacteriocins were low (below the threshold: 0) and the molecules were unstable as the instability index values were above the threshold of 40.
Furthermore, five proteins with analog structure with GT-1326 and GS-1368 are served in Table 3. These known proteins are useful to predict the function of the bacteriocins and its mechanism of action. Of five analog proteins found, four of them are functioned in encapsulation or compartment formation. To be more specific, the two highest TM-scores gained were the proteins with their function in iron encapsulation. Hence, it can be predicted that the role of the recent studied bacteriocins is in iron encapsulation process.
After obtaining the structure model as well as the physicochemical property data of bacteriocins, the in silico docking analysis was done to predict the interactions occurred between bacteriocins and putative target protein on P. larvae. The putative target protein was predicted based on the target protein of known bacteriocin with the same function as GT-1326 and GS-1368. Thus, lipoteichoic acid synthase was determined as it is the target of iron-chelating bacteriocin i.e., lactoferrin (Rosa et al., 2017). The in silico docking results between GT-1326 and lipoteichoic acid synthase are served in Fig. 4 in which the interactions among residues are depicted in Table 4.
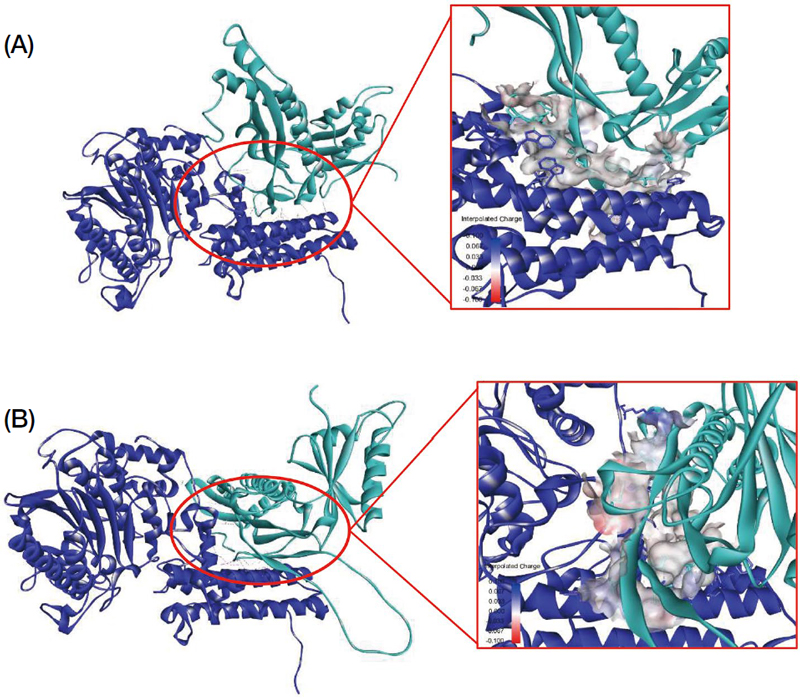
The results of in silico docking process between bacteriocins (greenish blue ribbon) and lipoteichoic acid synthase (dark-blue ribbon) through hydrogen bond (green dash line), electrostatic bonds (orange dash line), and hydrophobic bonds (purple dash line). (A) GT-1326 showed interactions with lipoteichoic acid synthase of P. larvae through three hydrogen bonds, one electrostatic bond, and 11 hydrophobic bonds; meanwhile, (B) GS-1368 interacted with lipoteichoic acid synthase of P. larvae through one hydrogen bond, two electrostatic bonds, and 15 hydrophobic bonds.
DISCUSSION
Bacteriocin has been widely known to be able to inhibit the growth of pathogenic bacteria. This antimicrobial agent is promising to develop as the potential benefits carried by bacteriocin along with their antibacterial activity. The bacteriocins studied in this research showed the potential as antibacterial agent against P. larvae. The extracted bacteriocins (i.e., GT-1326 and GS-1368) were purified and confirmed using SDS-PAGE separation, which then followed by visualization using coomasie brilliant blue (Fig. 1). The bands of purified bacteriocins were observed at identical position (approximately at 32 kDa). These results are in line with the ExPAsy prediction (Table 2). Therefore, it was proven that the two genes amplified and cloned are the bacteriocins coding genes.
After confirming bacteriocins’ physicochemical properties, the antimicrobial activities of GT-1326 and GS-1368 were tested using MIC method. The results (Table 1) showed that there were growth inhibition activities of both bacteriocins in which the MIC value was the same of 800 μg/mL. Albeit that the MIC results were not as effective as ampicillin (25 μg/mL), but the antimicrobial activity of the bacteriocins are need to be considered as the potential sources as a backbone in encountering bacteria, particularly P. larvae. There were also several previous reports which revealed the lower MIC values of bacteriocins compared to conventional antibiotics (Ma et al., 2022; Zhang et al., 2022).
Furthermore, the cell leakage assay was done by measuring cell free supernatant (CFS) of treated P. larvae in which the results are served in Fig. 2. Based on the figure, it was clear that the cell leakage occurred as the nucleic acid observed in the CFS at the wavelength 260 nm. The highest value observed in CFS of P. larvae treated using GT-1326 was about 0.007, while those which were treated using GS-1368 was approximately 0.017. This means that the nucleic acid concentrations of P. larvae CFS treated using GT-1326 and GS-1368 (without any dilution factor) were 0.35 and 0.85 μg/mL respectively. Hence, it is assumed that the mechanism of action of the bacteriocins is by disrupting bacterial cell membrane (Ma et al., 2022).
The findings of both MIC and cell leakage assays were supported by the results of bioinformatic analysis. Based on the molecule structures of GT-1326 and GS-1368 shown in Table 2, these two comprised of α chain with six α-helix structure. This α-helix structure is presumed to be the part of bacteriocins which are capable to form pores on P. larvae cell membrane (Vázquez et al., 2018). Contrarily, the inhibition ability was not strong which may be caused by the instability of the proteins were above the threshold. The values obtained were 50.61 for GT-1326 and 50.04 for GS-1368. Meanwhile, the instability index of stable molecule should not higher than 40. Thus, further research needs to be done to find the stabilization strategies (Akbarian and Chen, 2022).
To further investigation, molecule structure of the bacteriocin is important to predict its function. The two bacteriocins studied in this research are analog with the known protein in PDB, namely encapsulin (PDB ID: 6nj8) in which the function is as iron storage compartment. This protein is possessed by Quasibacillus thermotolerans (Giessen et al., 2019). The analog structure is predicted to perform the same function i.e., iron sequestration. GT-1326 and GS-1368 are predicted to bind to P. larvae surface receptor and catch iron. Thus, the lack-of-iron condition leads the death of P. larvae.
Bacteria should sequester iron as its essential demand of many metabolic processes (Nairz et al., 2010), including P. larvae (Grady et al., 2016). It is known that Paenibacillus show an ability in iron acquisition in several ways, one of which is by expressing siderophores (Grady et al., 2016). It was also revealed that iron sequestration protein functions as antibacterial agent. Mammals’ milk-contained protein, namely lactoferrin (Masson and Heremans, 1971) was reported to perform antimicrobial as well as anti-biofilm activity dependent on its iron-binding ability (Chandran et al., 2021). This protein interacts with lipopolysaccharide (of gram-negative bacteria) or lipoteichoic acid (of gram-positive bacteria) (Rosa et al., 2017).
Acknowledging this ability in iron-sequestration, an interaction between GT-1326 or GS-1368 and lipoteichoic acid of P. larvae were observed by performing in silico docking. The results (Table 4) revealed that there were three types of interactions occurred between bacteriocins and lipoteichoic acid. GT-1326 and lipoteichoic acid formed four hydrogen bonds, five electrostatic bonds as well as nine hydrophobic bonds. The main interaction occurred between bacteriocin and amino acid TYR41 of lipoteichoic acid of P. larvae. Meanwhile, GS-1368 and lipoteichoic acid formed one hydrogen bond, two electrostatic bonds, and 16 hydrophobic bonds, in which the main interaction occurred between bacteriocin and ASP 90 of lipoteichoic acid. An interesting fact of iron-chelating antimicrobial agent is that it also demonstrated bacteria killing independently from its iron-binding ability. It can compete to the host cells or to microbial surface components (Valenti and Antonini, 2005). In the other words, GT-1326 and GS-1368 interactions with receptor on P. larvae surface can either inhibit them to attach to the host cells and/or compete them in gaining iron needed.
Acknowledgments
This research was supported by the IPET (116094-03-1-SB010).
FOOTNOTES
Financial Disclosure: This research was conducted in the absence of any commercial or financial relationships that could be construed as posing potential conflicts of interest.
References
- AccessScience Editors. 2017. U.S. bans antibiotics use for enhancing growth in livestock. Editorial Briefing Jan- uary 2017. https://www.accessscience.com/content/briefing/aBR0125171, . Accessed 9 June. 2024.
-
Aizen, M. A., L. A. Garibaldi, S. A. Cunningham and A. M. Klein. 2008. Long-term global trends in crop yield and production reveal no current pollination shortage but increasing pollinator dependency. Curr. Biol. 18(20): 1572-1575.
[https://doi.org/10.1016/j.cub.2008.08.066]

-
Akbarian, M. and S. H. Chen. 2022. Instability challenges and stabilization strategies of pharmaceutical proteins. Phar- maceutics 14(11): 1-38.
[https://doi.org/10.3390/pharmaceutics14112533]

-
Ash, C., F. G. Priest and M. D. Collins. 1993. Molecular identifi- cation of rRNA group 3 bacilli (Ash, Farrow, Wallbanks and Collins) using a PCR probe test - Proposal for the creation of a new genus Paenibacillus. Antonie van Leeuwenhoek 64: 253-260.
[https://doi.org/10.1007/BF00873085]

- Chandran, D., A. U. Haq and S. S. Randhawa. 2021. Lactoferrin a potential therapeutic candidate. The Science World 1(4): 105-111.
- Dassault Systèmes, B. 2020. BIOVIA Discovery Studio Visual- izer (p. v21.1.0.20298). Dassault Systèmes.
-
De Graaf, D. C., A. M. Alippi, K. Antúnez, K. A. Aronstein, G. Budge, D. De Koker, L. De Smet, D. W. Dingman, J. D. Evans, L. J. Foster, A. Fünfhaus, E. Garcia-Gonzalez, A. Gregorc, H. Human, K. D. Murray, B. K. Nguyen, L. Poppinga, M. Spivak, D. Van Engelsdorp, W. Selwyn and E. Genersch. 2013. Standard methods for American foulbrood research. J. Apic. Res. 52(1): 1-27.
[https://doi.org/10.3896/IBRA.1.52.1.11]

-
Debeljak, Z. and D. Vidanovi. 2023. American Foulbrood - Old and always new challenge. Vet. Sci. 10(3): 180.
[https://doi.org/10.3390/vetsci10030180]

-
Fünfhaus, A., J. Göbel, J. Ebeling, H. Knispel, E. Garcia-Gon- zalez and E. Genersch. 2018. Swarming motility and biofilm formation of Paenibacillus larvae, the etiolog- ical agent of American Foulbrood of honey bees (Apis mellifera). Sci. Rep. 8: 8840.
[https://doi.org/10.1038/s41598-018-27193-8]

-
Genersch, E. 2010. American foulbrood in honeybees and its causative agent, Paenibacillus larvae. J. Invertebr. Pathol. 103(Suppl. 1): S10-S19.
[https://doi.org/10.1016/j.jip.2009.06.015]

-
Giessen, T. W., B. J. Orlando, A. A. Verdegaal, M. G. Chambers, J. Gardener, D. C. Bell, G. Birrane, M. Liao and P. A. Silver. 2019. Large protein organelles form a new iron sequestration system with high storage capacity. eLife 8: e46070.
[https://doi.org/10.7554/eLife.46070]

-
Grady, E. N., J. MacDonald, L. Liu, A. Richman and Z. C. Yuan. 2016. Current knowledge and perspectives of Paeni- bacillus: A review. Microb. Cell Fact. 15: 203.
[https://doi.org/10.1186/s12934-016-0603-7]

-
Hung, K. L. J., J. M. Kingston, M. Albrecht, D. A. Holway and J. R. Kohn. 2018. The worldwide importance of honey bees as pollinators in natural habitats. Proc. R. Soc. B. 285: 20172140.
[https://doi.org/10.1098/rspb.2017.2140]

-
Khanna, V., A. Jordan, H. M. Patch and C. M. Grozinger. 2021. Economic dependence and vulnerability of united states agricultural sector on insect-mediated pollination ser- vice. Environ. Sci. Technol. 55(4): 2243-2253.
[https://doi.org/10.1021/acs.est.0c04786]

- Laate, E. A., J. P. Emunu, A. Duering and L. Ovinge. 2020. Potential economic impact of European and American Foulbrrod on Alberta’s beekeeping industry. 5p. Govern- ment of Alberta, Alberta.
-
Ma, Y., Z. Guo, B. Xia, Y. Zhang, X. Liu, Y. Yu, N. Tang, X. Tong, M. Wang, X. Ye, J. Feng, Y. Chen and J. Wang. 2022. Identification of antimicrobial peptides from the human gut microbiome using deep learning. Nat. Bio- technol. 40(6): 921-931.
[https://doi.org/10.1038/s41587-022-01226-0]

-
Masson, P. L. and J. F. Heremans. 1971. Lactoferrin in milk from different species. Comp. Biochem. Physiol. B 39(1): 119-129.
[https://doi.org/10.1016/0305-0491(71)90258-6]

-
Nairz, M., A. Schroll, T. Sonnweber and G. Weiss. 2010. The struggle for iron - a metal at the host-pathogen interface. Cell Microbiol. 12(12): 1691-1702.
[https://doi.org/10.1111/j.1462-5822.2010.01529.x]

-
Okamoto, M., M. Kumagai, H. Kanamori and D. Takamatsu. 2021. Antimicrobial resistance genes in bacteria isolated from Japanese honey, and their potential for conferring macrolide and lincosamide resistance in the american foulbrood pathogen Paenibacillus larvae. Front Micro- biol. 12: 667096.
[https://doi.org/10.3389/fmicb.2021.667096]

-
Rosa, L., A. Cutone, M. S. Lepanto, R. Paesano and P. Valenti. 2017. Lactoferrin: A natural glycoprotein involved in iron and inflammatory homeostasis. Int. J. Mol. Sci. 18(9): 1985.
[https://doi.org/10.3390/ijms18091985]

-
Sarkar, T., M. Chetia and S. Chatterjee. 2021. Antimicrobial peptides and proteins: From nature’s reservoir to the lab- oratory and beyond. Front Chem. 9: 691532.
[https://doi.org/10.3389/fchem.2021.691532]

- Shahbandeh, M. 2022. Honey market worldwide and in the U.S. - statistics & facts. Dec 9. https://www.statista.com/topics/5090/honey-market-worldwide/#topicOverview/, . Acces- sed 27 July. 2023.
-
Tian, B., N. H. Fadhil, J. E. Powell, W. K. Kwong and N. A. Moran. 2012. Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio 3(6): e00377-12.
[https://doi.org/10.1128/mBio.00377-12]

-
Valenti, P. and G. Antonini. 2005. Lactoferrin: An important host defence against microbial and viral attack. Cell. Mol. Life Sci. 62(22): 2576-2587.
[https://doi.org/10.1007/s00018-005-5372-0]

- Vázquez, A., R. Perdomo-Morales and V. Montero-Alejo. 2018. Natural antimicrobial peptides. Biotecnol. Apl. 35(4): 4101-4107.
- World Animal Protection. 2022. January 2022. EU bans the routine use of antibiotics in farmed animals. January 8. https://www.worldanimalprotection.org/latest/news/eu-bans-antibiotic-overuse-farmed-animals/, . Accessed 9 June. 2024.
-
Zebrowska, J., M. Witkowska, A. Struck, P. E. Laszuk, E. Rac- zuk, M. Ponikowska, P. M. Skowron and A. Zylicz-Sta- chula. 2022. Antimicrobial potential of the Genera Geo- bacillus and Parageobacillus, as well as endolysins bio- synthesized by their pacteriophages. Antibiotics (Basel) 11(2): 242.
[https://doi.org/10.3390/antibiotics11020242]

-
Zhang, J., S. Gu, T. Zhang, Y. Wu, J. Ma, L. Zhao, X. Li and J. Zhang. 2022. Characterization and antibacterial modes of action of bacteriocins from Bacillus coagulans CGMCC 9951 against Listeria monocytogenes. LWT-Food Sci. Technol. 160: 113272.
[https://doi.org/10.1016/j.lwt.2022.113272]