Rapid Diagnostic Method of Deformed Wing Virus Using Modified Reverse Transcription Recombinase-polymerase Amplification Method
Abstract
Isothermal amplification methods have become a cornerstone for pathogen detection, offering a simplified alternative to the polymerase chain reaction by enabling target gene amplification at a steady temperature. This study focuses on recombinase polymerase amplification (RPA), an isothermal technique capable of amplifying a target gene within 20 to 30 minutes at a constant 37°C. Leveraging this approach, we sought to devise a rapid and precise diagnostic method for the deformed wing virus (DWV) in honey bees. By incorporating reverse transcriptase into the TwistAmp® Basic kit, we facilitated one-step RT-RPA for DWV detection. Prior to this, we meticulously designed a suitable primer set and fine-tuned the reaction conditions, varying temperature, duration, primer selection, and magnesium ion concentration to optimize the process. Our newly developed diagnostic method streamlines nucleic acid separation by utilizing the Quick ExtractTM DNA Extraction Solution. As a result, the entire process-from extraction to diagnosis-now takes just 70 minutes. Moreover, this method’s simplicity enables on-site field diagnosis, enhancing its practical application for immediate DWV detection in honey bees.
Keywords:
Honey bee, DWV, Isothermal amplification method, RT-RPA, Quick RNA extractionINTRODUCTION
Honey bees are invaluable not only as honey producers but also as pollinators, contributing to approximately 50% of the pollination required for annual crop production (Jung, 2008). Economically, this represents a staggering 5.9 trillion won, nearly 18 times the domestic beekeeping industry’s primary production value of 260 million U.S. dollars (Jung, 2008). Since the initial identification of a honey bee-infecting virus in 1913 (Grozinger and Flenniken, 2019), a multitude of viral threats have emerged, posing significant risks to the beekeeping sector.
Among these, the deformed wing virus (DWV), a causative agent of wing deformity disease, is a notable single-stranded RNA virus belonging to the Iflaviridae family (Jeong, 2012). Initially discovered in asympto- matic adult honeybees in Egypt in 1977 and termed Egyptian bee virus (Bailey et al., 1979), it was later designated as wing fever virus in 1982 following the identification of characteristic symptoms in Japan (De Miranda and Genersch, 2010).
In South Korea, DWV was first reported in Yeoju, Gyeonggi-do, in 2005 (Lee et al., 2005). Subsequently, it has proliferated across most regions of South Korea, with a marked increase in infection rates. In the first half of 2022, DWV exhibited the highest infection rate among 14 honey bee diseases (Kim and Park, 2022) and is also implicated in colony collapse disorder (De Miranda and Genersch, 2010; Martin and Brettell, 2019). The association between DWV infection and Varroa mite infestation is well-documented (De Miranda and Genersch, 2010), with evidence of horizontal transmission (Choi et al., 2008a). The virus primarily manifests as wing deformities in young adult honey bees, along with abdominal abnormalities, discoloration, and paralysis (Jeong, 2012).
The polymerase chain reaction (PCR) has been the standard diagnostic tool for honey bee viruses (Bakonyi et al., 2002; Chen et al., 2004, 2005; Choi et al., 2008a, 2008b), due to its high accuracy and sensitivity (Yue and Genersch, 2005; Yoo et al., 2007). However, PCR requires specialized equipment and extended diagnosis times (Piepenburg et al., 2006), prompting the need for alternative methods.
Isothermal amplification techniques, such as rolling circle amplification (RCA), loop-mediated isothermal amplification (LAMP), and recombinase polymerase amplification (RPA), offer promising alternatives for pathogen detection (Zhao et al., 2015). RPA, in particular, facilitates target gene sequence amplification under constant temperature conditions, utilizing recombinase uvsX, co-factor uvsY, and Sau polymerase for DNA elongation (Piepenburg et al., 2006; Min et al., 2016b). Recommended to be conducted at 37-42℃, the RPA reaction typically completes within 20 minutes (Subbotin, 2019). Its efficacy as a diagnostic method has been demonstrated in recent studies, including pathogen detection attempts for black queen cell virus (BQCV) (Min et al., 2016a). Until recently, diagnosis through RPA has been attempted for Black queen cell virus (BQCV) (Lim et al., 2016) and sacbrood virus (SBV) (Min et al., 2016a), though not yet for DWV.
This research endeavors to establish a swift RT-RPA diagnostic technique for DWV in honey bees, comparing the diagnostic duration against PCR and exploring various RNA extraction methods to potentially expedite DWV detection.
MATERIALS AND METHODS
1. Collection of honey bee samples
On July 12, 2021, adult honey bees (Apis mellifera) exhibiting symptoms suggestive of DWV infection were collected from an apiary at the Insect Ecology-Pollinology Laboratory, Andong National University. Honey bees displaying external abnormalities, such as shortened or malformed wings and abdomens, were categorized into groups based on their relative activity levels and colony size. If the number of honey bees in a colony was more than 40,000, it was designated as ‘strong’, and if the number of bees in a colony was less than 20,000, it was designated as ‘weak’.
2. DWV detection via PCR and sequence analysis
For each mentioned group, two bees were randomly selected and then homogenized using a mortar and pestle. Viral RNA extraction was performed using the Viral Gene-spinTM Viral DNA/RNA Extraction Kit (Intron Biotechnology, Seongnam, Korea), following the manufacturer’s instructions. The process involved the addition of 500 μL of lysis buffer to the sample, incubation at 65℃ for 10 minutes, and subsequent separation of 250 μL of supernatant. This was followed by the addition of 350 μL of binding buffer, careful mixing, and centrifugation using the provided spin column. The column was washed with 500 μL of washing buffer A, then with 500 μL of washing buffer B, and finally centrifuged without additives to dry the column. RNA was eluted with 40 μL of elution buffer, incubated at room temperature (20-25℃) for 5 minutes, and centrifuged to collect the RNA, which was stored at -80℃ until further use.
Primers for DWV diagnosis were ordered based on previous studies (Lee et al., 2018) (Table 1) and synthesized by Macrogen (Seoul, Korea). One-Step RT-PCR was conducted using these primers and SuPrimeScript RT-PCR Premix (2×) (GENETBIO, Daejeon, Korea), negating the need for a separate cDNA synthesis step. To prepare the mixture, 10 μL of premix, 7 μL of nuclease-free water, and 1 μL each of forward primer and reverse primer were mixed. Afterwards, 1 μL of the extracted DNA template was added and PCR was performed with a total of 20 μL of the mixture for each sample. The protocol included an initial cDNA synthesis at 50℃ for 30 minutes, followed by initial denaturation at 95℃ for 5 minutes, 35 cycles of denaturation at 95℃ for 30 seconds, annealing at 55℃ for 30 seconds, extension at 72℃ for 1 minute, and a final extension at 72℃ for 5 minutes. The amplified products were sequenced by Macrogen after agarose gel electrophoresis and purification with AccuPrep® PCR/Gel Purification Kit (Bioneer, Daejeon, Korea).
3. Optimization of RPA Diagnostic Conditions for DWV
Primer sets for RPA were designed using Primer-BLAST (NCBI, Bethesda, MD, USA), adhering to guidelines recommending a product length of 100-200 bp, primer lengths of 20-35 nucleotides, and a GC content of 40-60% (Daher et al., 2016). Five primer sets were tested using the TwistAmp® Basic Kit (Twist-Dx, Maidenhead, United Kingdom). The reaction mix included 2.1 μL of each primer at 10 μM, 29.5 μL of rehydration buffer, 10.3 μL of nuclease-free water, 3 μL of RNA template, and 0.5 μL of RevertAid Reverse Transcriptase (Thermo Fisher Scientific, Waltham, MA, USA). The addition of 2.5 μL of 280 mM Magnesium Acetate (MgOAc) initiated the RT-RPA reaction at 37℃ for 35 minutes. The resulting products were analyzed via 2% agarose gel electrophoresis to select the most effective primer set.
The optimal reaction temperature for RPA was determined by testing the reaction mix at various temperatures ranging from 30.8 to 50.8℃, using the T100 Thermal Cycler (Bio-Rad, Hercules, California, USA). The clearest amplification results dictated the selection of the ideal temperature.
To ascertain the most effective reaction time, the reaction mixtures, excluding MgOAc, were prepared and stored on ice. MgOAc was added at 5-minute intervals, and the reaction was allowed to proceed at the optimal temperature. Reaction times varied from 0 to 50 minutes, with the samples subsequently chilled to halt further reactions. Gel electrophoresis was used to evaluate the clarity of the amplification product across different reaction times.
The concentration of primers was also varied from the recommended 2.1 μL to 0, 0.5, 1, 1.5, and 2 times the recommended amount, with the reaction volume adjusted to 50 μL by altering the amount of nuclease-free water accordingly. Post-reaction, samples were chilled, and gel electrophoresis was performed to assess the clarity of the amplification products relative to primer concentration.
Finally, the amount of MgOAc, crucial for the RPA reaction, was adjusted from the recommended 2.5 μL to 0, 0.5, 1, 1.5, and 2 times the recommended amount. The optimal primer and MgOAc concentrations, as determined from previous experiments, were used to induce the reaction under the ideal temperature and time conditions. The clarity of the amplification product in relation to the MgOAc concentration was compared through gel electrophoresis.
4. Quick nucleic acid extraction method
Unlike existing DNA/RNA extraction methods, there is a method that can extract DNA/RNA in one step without filtration. A technique for diagnosing diseases by extracting RNA using Quick ExtractTM DNA Extraction Solution (LGC, Teddington, United Kingdom) and then performing RT-PCR was recently presented (Ladha et al., 2020). When extracting DNA/RNA through this method, it is possible to extract it with a reaction time of only 5 minutes. In addition, the time required is reduced by 1/3 compared to using Viral DNA/RNA Extraction Kit (Intron Biotechnology) using the existing extraction method (Ladha et al., 2020). Therefore, RNA was extracted using the quick extraction buffer, and the process was followed. First, the bees are pulverized and then mixed with nuclease-free water to produce a pulverization liquid. Next, mix the grinding liquid and DNA extraction solution in equal proportions, 200 μL each. After maintaining the temperature at 95℃ for 5 minutes, centrifuge at 13,000 rpm for 1 minute and extract the supernatant to confirm whether DWV can be diagnosed through RT-PCR and RT-RPA described above. For comparison, existing viral DNA/RNA Extraction used results were compared with the diagnostic results used.
RESULTS
1. Confirmation of DWV infection
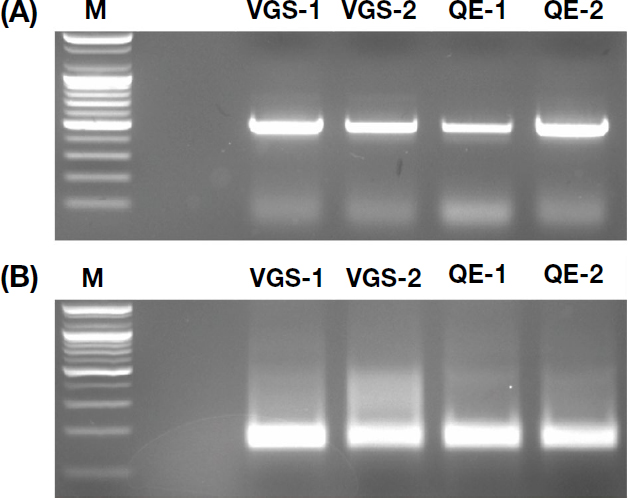
RT-PCR analysis of honey bee samples detected a 479 bp amplicon corresponding to the expected size for DWV (Fig. 1). Sequence alignment against NCBI Gen-Bank database (BLASTn) revealed a 96.95% similarity with the sequence previously identified in South Korea (JX878305), indicating significant sequence conservation (Fig. 2).

Electrophoretic analysis of RT-PCR products for DWV detection in honeybees. The gel shows bands for the stronger group (S), the weaker group (W), a 100 bp DNA Ladder (M), and a no-template control (N).
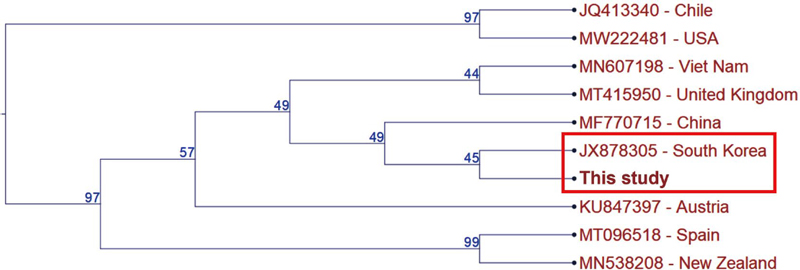
Phylogenetic tree depicting the genetic relationship of DWV sequences. The tree includes sequences from South Korea (Accession No. JX878305), the United States (MW222481), New Zealand (MN538208), Spain (MT096518), Chile (JQ413340), Austria (KU847397), China (MF770715), the United Kingdom (MT415950), and Vietnam (MN607198).
2. Optimized RPA diagnostic conditions for DWV
Electrophoretic analysis of RT-RPA products, using five custom-designed primer pairs, resulted in visible amplification across all sets (Fig. 3). Primer pairs 1 through 3 showed no significant differences in band intensity or clarity; thus, pair 1 was selected for further experimentation due to its ease of use.
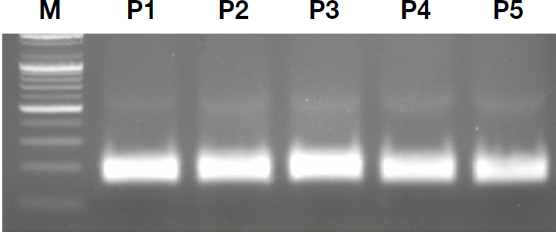
Primer efficacy for RT-RPA optimization. The gel compares amplification results using five different primer pairs, labeled P1 through P5.
Temperature optimization experiments, conducted in ranges of 40.8-50.8℃ and 30.8-40.8℃, revealed that amplification was successful at temperatures up to 44.7℃ but not at higher temperatures (Fig. 4A). In the lower temperature range, clear amplification was observed starting from 37℃ (Fig. 4B). Further refinement identified 41℃ as the optimal temperature, providing the clearest amplification product within the effective range of 35-45℃ (Fig. 4C).
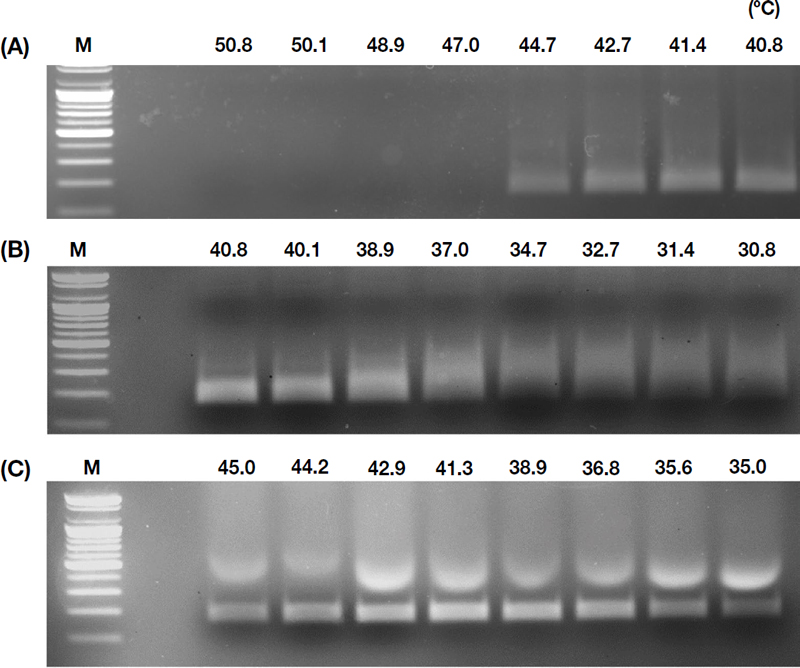
Temperature optimization for RT-RPA. (A) compares results at temperatures ranging from 40.8 to 50.8℃, (B) from 30.8 to 40.8℃, and (C) within the refined effective range of 35.0 to 45.0℃.
Time optimization studies indicated that amplification was not detectable immediately after MgOAc addition but became visible after 10 minutes, with the most distinct results appearing after 25 minutes (Fig. 5). Considering field sensitivity requirements, a 25-minute reaction at 41℃ was deemed optimal for diagnosis.
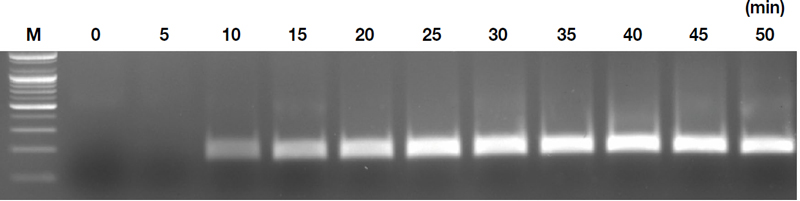
Time optimization for RT-RPA. The gel compares amplification results at 5-minute intervals under an optimal temperature of 41.0℃.
Primer concentration experiments showed that while a 4.20 μL addition produced the most intense bands, it also caused excessive spreading, potentially complicating diagnostic decisions (Fig. 6A). A primer addition of 3.15 μL for both forward and reverse primers was found to be most suitable for clear and interpretable results.
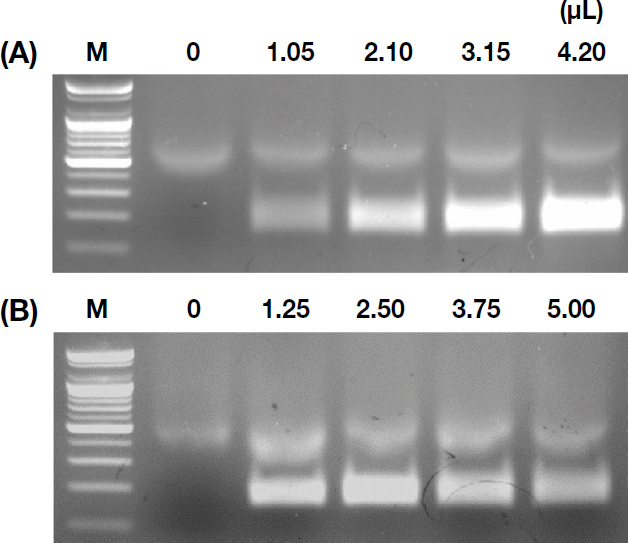
Optimization of RT-RPA based on reagent concentrations. (A) The impact of varying primer concentrations from 0 × to 2 ×. (B) The effects of different MgOAc concentrations.
MgOAc concentration trials concluded that 2.5 μL provided the clearest amplification, with higher concentrations resulting in fainter bands (Fig. 6B).
3. Comparison of nucleic acid extraction methods for DWV detection
We extracted RNA from honey bee samples using Quick ExtractTM DNA Extraction Solution and Viral Gene-spinTM Viral DNA/RNA Extraction Kit and performed RT-PCR. Agarose gel electrophoresis revealed consistent amplification products targeting DWV in both cases (Fig. 7A). Similarly, RT-RPA using the same templates confirmed specific DWV amplification products (Fig. 7B). The results obtained with the simplified extraction method were comparable to those from the existing diagnostic approach.
DISCUSSION
In this study, we developed a streamlined diagnostic method for detecting DWV using RT-RPA. Our approach significantly reduces the overall reaction time compared to the existing method. RNA extraction using Quick ExtractTM DNA Extraction Solution took only 5 minutes. Optimized reaction mixture preparation involved specific primer pairs (3.15 μL each), rehydration buffer (29.5 μL), reverse transcriptase (0.5 μL), nuclease-free water (5.2 μL), and RNA template (6 μL). Addition of 2.5 μL MgOAc followed by incubation at 41℃ for 25 minutes completed the reaction setup. The clearest results were obtained when electrophoresis was performed on a 2% agarose gel for 40 minutes. The conventional diagnostic method involves RNA extraction (40 minutes), RT-PCR amplification (130 minutes), and electrophoresis on a 1% agarose gel (20 minutes), totaling 190 minutes. Our newly designed method reduces the reaction time to just 70 minutes, saving approximately 120 minutes. In summary, our optimized protocol offers a rapid and efficient approach for DWV diagnosis.
We have developed a novel diagnostic method for DWV utilizing RNA-specific RPA. RPA is increasingly favored for pathogen diagnosis due to its rapid reaction time and comparable, if not superior, sensitivity to PCR under isothermal conditions. The integration of the CRISPR system with RPA further underscores the potential of this approach for viral diagnostics (Tan et al., 2022). Our method leverages precedents that have validated its application to virus detection by diagnosing Korean cyst virus in honey bees (Min et al., 2016a) and cymbidium mosaic virus from orchid plants.
RPA has several advantages in diagnosing pathogens. It is highly sensitive and reacts specifically, and has a wide temperature range effective for isothermal reaction, so there is no need for complex thermal change treatment or a thermocycler (Daher et al., 2016). However, due to high sensitivity, non-specific amplicons may occur (Li et al., 2019). To solve this problem, similar to designing specific primers for PCR diagnostics, the generation of non-specific amplicons can be reduced by designing primers from the most conserved sequences of the pathogen and performing the reverse transcription and amplification steps in one step (Tan et al., 2022). Additionally, as shown in the results of this study, it has the advantage of taking a much shorter time for diagnosis than PCR.
In this study, we conducted a comparative analysis of two nucleic acid extraction methods aimed at optimizing the extraction process while preserving diagnostic accuracy. Various RNA extraction techniques are employed in diagnostic settings, each with distinct strengths and limitations. Silica column-based RNA extraction kits are widely favored in laboratories due to their straightforward protocols. The methods evaluated in our study included a simplified buffer-based approach using Quick ExtractTM DNA Extraction Solution and a conventional method employing the Viral Gene-spinTM Viral DNA/RNA Extraction Kit. Our objective was to assess whether the streamlined extraction process with Quick ExtractTM maintained diagnostic accuracy comparable to the Viral Gene-spinTM method. Our results indicated no significant differences between the two methods, suggesting that the quicker nucleic acid separation facilitated by Quick ExtractTM does not compromise the reliability of RT-RPA assays. These findings underscore the potential of simplified extraction protocols in enhancing efficiency without sacrificing diagnostic robustness.
In summary, our approach successfully achieves isothermal amplification of DWV’s target gene via RPA, reducing the diagnosis time to 70 minutes, a significant improvement over the 190 minutes required by conventional methods. This expedited process, coupled with the reduced equipment needs, facilitates on-site diagnosis. Looking forward, we plan to commercialize this method and expand its application to the diagnosis of various honey bee viruses.
References
-
Bailey, L., J. Carpenter and R. Woods. 1979. Egypt bee virus and Australian isolates of Kashmir bee virus. J. Gen. Virol. 43(3): 641-647.
[https://doi.org/10.1099/0022-1317-43-3-641]

-
Bakonyi, T., R. Farkas, A. Szendröi, M. Dobos-Kovács and M. Rusvai. 2002. Detection of acute bee paralysis virus by RT-PCR in honey bee and Varroa destructor field sam- ples: rapid screening of representative Hungarian apiar- ies. Apidologie 33(1): 63-74.
[https://doi.org/10.1051/apido:2001004]

-
Chen, Y., J. S. Pettis and M. F. Feldlaufer. 2005. Detection of multiple viruses in queens of the honey bee Apis melli- fera L. J. Invertebr. Pathol. 90(2): 118-121.
[https://doi.org/10.1016/j.jip.2005.08.005]

-
Chen, Y., Y. Zhao, J. Hammond, H.-T. Hsu, J. Evans and M. Feldlaufer. 2004. Multiple virus infections in the honey bee and genome divergence of honey bee viruses. J. Invertebr. Pathol. 87(2-3): 84-93.
[https://doi.org/10.1016/j.jip.2004.07.005]

- Choi, Y.-S., M.-L. Le, M.-Y. Le and K.-G. Lee. 2008a. Occur- rence of seven honeybee viruses and researchof disease occurrence in Korean apiaries. Korean J. Apic. 23(2): 153-159.
- Choi, Y.-S., M.-L. Le, M.-Y. Le and K.-G. Lee. 2008b. Detection of seven bee viruses from Varroa destructor mite. Korean J. Apic. 23(3): 171-176.
-
Daher, R. K., G. Stewart, M. Boissinot and M. G. Bergeron. 2016. Recombinase polymerase amplification for diag- nostic applications. Clin. Chem. 62(7): 947-958.
[https://doi.org/10.1373/clinchem.2015.245829]

-
De Miranda, J. R. and E. Genersch. 2010. Deformed wing virus. J. Invertebr. Pathol. 103: S48-S61.
[https://doi.org/10.1016/j.jip.2009.06.012]

-
Grozinger, C. M. and M. L. Flenniken. 2019. Bee viruses: Ecol- ogy, pathogenicity, and impacts. Annu. Rev. Entomol. 64: 205-226.
[https://doi.org/10.1146/annurev-ento-011118-111942]

- Jeong, N.-G. 2012. Diseases on Honeybee (Virus)-1. J. Kor. Vet. Med. Ass. 48(12): 748-762.
- Jung, C. 2008. Economic value of honeybee pollination on major fruit and vegetable crops in Korea. Korean J. Apic. 2: 147-152.
-
Kim, K.-Y. and Y.-S. Park. 2022. Bee epidemics and trends in treatment technology development. Korean J. Apic. 37(3): 315-329.
[https://doi.org/10.17519/apiculture.2022.09.37.3.315]

-
Ladha, A., J. Joung, O. O. Abudayyeh, J. S. Gootenberg and F. Zhang. 2020. A 5-min RNA preparation method for COVID-19 detection with RT-qPCR. MedRxiv 20200323: 1-3.
[https://doi.org/10.1101/2020.05.07.20055947]

- Lee, H.-M., D.-B. Lee, S.-H. Han, M.-L. Lee, Y.-K. Lim and B.-S. Yoon. 2005. Identification of deformed wing virus from the honeybee in Korea and establishmrnt of PCR detec- tion method. Korean J. Apic. 20(2): 85-94.
- Lee, I.-H., J.-Y. Kim, J.-W. Choi, K. Ba-Ra-Da, B.-R. Jung, Park, J.-S., H.-M. Na and Y.-H. Kim. 2018. Prevalence of honeybee (Apis mellifera) diseases in Gwangju. Korean J. Vet. Serv. 41(2): 111-118.
-
Li, J., J. Macdonald and F. Von Stetten. 2019. A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 144(1): 31-67.
[https://doi.org/10.1039/C8AN01621F]

-
Lim, S.-J., G. T. H. Luong, J. H. Wang, S. H. Min and B.-S. Yoon. 2016. Rapid detection of black queen cell virus from honeybee by real-time recombinase polymerase amplification. Korean J. Apic. 31: 31.
[https://doi.org/10.17519/apiculture.2016.04.31.1.41]

-
Martin, S. J. and L. E. Brettell. 2019. Deformed wing virus in honeybees and other insects. Annu. Rev. Virol. 6(1): 49-69.
[https://doi.org/10.1146/annurev-virology-092818-015700]

-
Min, S.-H., J.-H. Wang, S.-J. Lim and B.-S. Yoon. 2016a. De- velopment of detection-method against Korean sacbrood virus (kSBV) using real-time sothermal recombinase polymerase amplification (RPA). Korean J. Apic. 31: 63.
[https://doi.org/10.17519/apiculture.2016.06.31.2.147]

-
Min, S.-H., J.-H. Wang, S.-J. Lim, C.-W. Lee and B.-S. Yoon. 2016b. Development of quantitative real-time recombi- nase polymerase amplification (qRT-RPA) method for quantitative detection against pathogenic virus in honey- bee. Korean J. Apic. 31: 147-156.
[https://doi.org/10.17519/apiculture.2016.06.31.2.147]

-
Piepenburg, O., C. H. Williams, D. L. Stemple and N. A. Armes. 2006. DNA detection using recombination proteins. PLOS Biol. 4(7): e204.
[https://doi.org/10.1371/journal.pbio.0040204]

-
Subbotin, S. A. 2019. Recombinase polymerase amplification assay for rapid detection of the root-knot nematode Meloidogyne enterolobii. Nematology 21(3): 243-251.
[https://doi.org/10.1163/15685411-00003210]

-
Tan, M., C. Liao, L. Liang, X. Yi, Z. Zhou and G. Wei. 2022. Recent advances in recombinase polymerase amplifica- tion: Principle, advantages, disadvantages and applica- tions. Front. Cell. Infect. Microbiol. 12: 1744.
[https://doi.org/10.3389/fcimb.2022.1019071]

- Yoo, M.-S., D.-W. Lee, I.-W. Kim, D.-S. Kim, S.-H. Kwon, Y.-G. Lim and B.-S. Yoon. 2007. Identification of black queen cell virus from the honeybee in Korea. Korean J. Apic. 22(1): 43-52.
-
Yue, C. and E. Genersch. 2005. RT-PCR analysis of Deformed wing virus in honeybees (Apis mellifera) and mites (Var- roa destructor). J. Gen. Virol. 86(12): 3419-3424.
[https://doi.org/10.1099/vir.0.81401-0]

-
Zhao, Y., F. Chen, Q. Li, L. Wang and C. Fan. 2015. Isothermal amplification of nucleic acids. Chem. Rev. 115(22): 12491-12545.
[https://doi.org/10.1021/acs.chemrev.5b00428]