
Differential Expression of Immune-Related Genes in Larval and Adult Apis mellifera Honeybees Challenged with Korean Sacbrood Virus
Abstract
Recently, the Korean sacbrood virus (KSBV) caused great loss of honeybee colonies in Korean Apis mellifera. Three antimicrobial peptide genes (AMPs) (abaecin (ABA), defensin (DEF) and hymenoptaecin (HYM)) and two immune-related enzyme genes (phenol-oxidase (PO) and lysozyme (LYS)) were investigated using qRT-PCR as a measure of the defense reactions of honeybee larvae and adults challenged with KSBV in vitro. Transcript levels of three AMPs were upregulated in adults exposed to KSBV, compared to control adults. ABA and DEF transcripts were 22.7 and 21.9 fold higher (P<0.05), respectively, after 24h exposure while HYM transcript did not change significantly. At this time point, PO and LYS mRNA levels were 18.08 and 16.03 fold higher, respectively, in KSBVinfected adult bees compared to control adults (P<0.05). By contrast, the larvae infected with KSBV had downregulated expression of AMPs and slight up-regulation of PO and LYS compared to the controls. These results suggest that the differential expression of the genes studied could indicate their involvement in defense processes against KSBV in adult and larva bees.
Keywords:
Honeybees, Apis mellifera (A. mellifera), Korean sacbrood virus (KSBV), Immune-related genes, Antimicrobial peptide genes (AMPs)INTRODUCTION
The honeybee Apis mellifera (A. mellifera) is an important insect economically, not only for honey production, but also for crop pollination. They are highly successful eusocial insects, with colonies that consist of a single queen, up to 80,000 sterile female workers, and male drones. Their social lifestyle and very high population density within their hives render honeybees especially vulnerable to infection by pathogens such as viruses, bacteria, fungi and parasites (protozoa, mites, flies, beetles and nematodes) (Barley and Ball, 1991; Evans and Pettis, 2005). To combat the increased risk of disease, honeybees have evolved novel behavioral, physiological and organizational adaptations to eliminate pathogens.
Honeybees, like all insects, lack an adaptive immune system to help combat infection by pathogens; hence, they rely solely on innate immune reactions based on a constitutively active cellular and an inducible humoral immune response (Hoffmann, 2003). Cellular immunity involves processes such as coagulation, phagocytosis, nodulation and encapsulation. Both nodulation and encapsulation are often accompanied by melanization, which is catalyzed by the PO. PO is thought to be a key component of the primary immune response of arthropods (Decker and Jaenicke, 2004). Melanization mediated by PO plays a major role in the immune defense of insect. LYS is another important enzyme in the insect immune response because it degrades Gram-positive and Gramnegative bacteria, and can promote the expression of other AMPs (Imler and Bulet, 2005). Alternatively, humoral immunity, which involves the synthesis of AMPs in response to infection by bacteria, fungi or parasites, requires stimulation by specific pathogen recognition. Numerous AMPs have now been identified in invertebrates, and are recognized as playing a key role in protection from pathogenic organisms. Among them, namely ABA (Casteels et al., 1990), HYM (Casteels et al., 1993) and DEF (Casteels - Josson et al., 1994) have been investigated extensively in the honeybee. All five ABA, HYM, DEF, PO and LYS are known to be involved in the immune defense reaction of bees directed against bacteria (Gatchenberger et al., 2012; Laughton et al., 2011; Randolt et al., 2008; Evans et al., 2004), but their effect on the immune response of A. mellifera to viruses is unknown (Azzami et al., 2012).
SBV is widely distributed on every continent and is one of about 18 bee viruses identified so far (Allen and Ball, 1996; Chen and Sieder, 2007; De Miranda et al., 2013). It attacks both the larvae and the adults of A. mellifera colonies, causing sacbrood disease (Ball, 1999) and serious damage to the colonies. In 2009, there was an outbreak of sacbrood in Korea, from which Korean SBV (KSBV) was isolated. Although KSBV shows a high homology with other SBV strains, it has unique motifs and causes different symptoms. KSBV generally referred to as a picorna-like virus, has a single-stranded positive-sense polyadenylated RNA genome of 8792 nucleotides (Choe et al., 2012). Today, SBV is the most common viral disease of bees reported but little is known genetic differences that could account for variations in honeybee tolerance to SBV. Moreover, behavioral differences between the honeybee brood and adults are likely to play an important role in disease susceptibility to different pathogens.
In this study, we have examined the expression of five immune-related genes as a measure of the defense reactions of A. mellifera worker bee larvae and adults challenged with KSBV, with the aim of identifying genes likely to be involved in the response to KSBV infection. The expression of five candidate genes, three encoding AMPs (ABA, DEF, and HYM) and two encoding immune- related enzymes (PO and LYS) were measured by qRTPCR. Although viruses do not possess peptidoglycan cellular membrane, known to be bacterial stimulus for the AMP response, we did find a difference in the mRNA expression levels of AMPs between control and KSBVinfected adult bees.
MATERIALS AND METHODS
In vitro rearing of worker bee larvae and adults
Worker bee larvae and adults were obtained from healthy colonies of A. mellifera in the apiary of the Parasite and Insect Disease Laboratory, Animal and Plant Quarantine Agency, Korea. All larvae and adults used were previously identified the absence of infection with any pathogen by RT-PCR. Newly hatched larvae were collected from combs with a special grafting tool and transferred to 24-well tissue culture plate filled with 50μl of basic diet. The daily amount and composition was prepared according to Peng et al. (1991) and Aupinel et al. (2005). It consisted of 6% (w/v) glucose, 6% (w/v) fructose, 1% (w/v) yeast extract and 50% (w/v) royal jelly (diet is fed to larvae on day). The grafted larvae collected from this colony were maintained at 35°C in a high (80%) humidity. Each day they were transferred to new tissue culture plates filled with fresh pre-warmed diet. Newly emerged free disease drones (up to 24 h old) were selected from the combs and divided into groups of ten in small plastic cages (12cm×6cm×3cm) for each series of experiments. These cages were incubated at 35°C and 80% humidity. Worker bee adults were supplied with a diet of 50% (w/v) sugar solution.
Preparation of KSBV suspensions
Honey bee larvae infected with KSBV were obtained from the local apiary in Gyeongsangbuk-do, South Korea. These larvae were ground in phosphate buffered saline (pH 7.4), then filtered first with a 0.45μm and then a 0.2μm Minisart filter (Sartorius, Gottingen, Germany). The concentration of KSBV in the suspension was estimated by real time-PCR.
Experimental KSBV infection
Larvae (3-4 days old) and young workers (1 day old) were artificially infected using the KSBV suspension. After testing various concentrations of the KSBV suspension, a concentration of 105-6 copies mixed directly with the feeding solution was used for inoculation; controls received liquid diet alone. All cages were maintained in an incubator at 35°C and 80% humidity. Each series of experiments with larvae and adults consisted of five groups and uninfected controls. After exposure to KSBV, five to ten individuals per group were collected 3, 6, 12, 24 and 48 h later and directly frozen at -80°C.
RNA extraction
Total RNA was extracted from each individual larva and adult using Viral Gene-spin Viral DNA/RNA extraction Kit (iNtRON Biotechnology, Sungnam, Korea) according to the manufacturer’s instructions. After removing DNA, the total RNA concentration was quantitated using an ultraviolet spectrophotometer (Thermo Scientific, USA) and stored at -80°C before use.
Genes and primers
To evaluate the immune response of honeybees with KSBV infection, the transcript levels of the genes encoding AMPs: ABA, HYM, DEF and immune-related enzymes: PO and LYS were assessed using primers previously described (Evans, 2006a; Yang and Cox-Foster, 2005). The transcript levels of β-actin, a moderately expressed housekeeping gene was used to normalize for variation in cDNA levels (Evans, 2006a). The primers were synthesized by Bioneer Corporation Ltd. (Daejeon, Korea). Table 1 summarizes the primers used.
Real-time PCR
Real-time PCR was performed in 8-tube strips (Bio-Rad, USA) and carried out using a CFX Connect Real-Time System (Bio-Rad, USA) and One Step SYBR PrimeScript RT-PCR Kit II (Perfect Real Time, TAKARA BIO, Japan), according to the manufacturer’s instructions. The reaction mixture with total volume of 20μl contained 10μl of 2x One Step SYBR RT-PCR Buffer IV, 0.4μM of each primer (Table 1), 0.8μl of PrimeScript 1 step enzyme Mix II, RNase free dH2O and 2μl of 50ng/μl RNA. The cycling program was as follows: an initial step at 42°C for 50 min for cDNA synthesis, then 95°C for 10 min followed by 40 cycles of 95°C for 30 sec, 59°C for 30 sec and 72°C for 30 sec. Fluorescence was measured in the annealing step. The specificity of the reaction was checked by analysis of the melting curve of the final amplified product (from 65°C to 95°C).
Normalization of the Real-time PCR data and statistical analysis
The amplification results for the different genes were represented by the threshold cycle (Ct) value, which is the number of cycles required to generate a fluorescent signal greater than a predefined threshold. mRNA levels were calculated relative to β-actin expression using the Bio-Rad CFX Manager Software (version 3.0). Differences in gene expression levels among the treatment groups for each gene of interest were analyzed using one-way ANOVA. P<0.05 was defined as statistically significant. Data are presented for overall mean transcript levels across all treatments.
RESULTS
The expression levels of ABA, HYM, DEF, PO and LYS genes were quantified by qRT-PCR in larva and adult A. mellifera worker bees after infection with KSBV for various lengths of time. In all cases, expression levels are represented by the mean of three measurements.
Expression level of three AMPs genes in honeybees upon KSBV infection
The mRNA levels of three AMPs genes were determined in worker honeybees 3, 6, 12, 24 and 48 h following inoculation with KSBV and compared to the levels in control worker bees at each time points. Transcript levels for ABA and DEF increased significantly upon exposure of adult bees to KSBV (Figs. 1A, B and C). ABA and HYM, transcript levels began increasing at 6 h and DEF at 3 h following infection. In seven independent inoculation experiments, after 24 h exposure, the ABA and DEF transcript levels were 22.7 and 21.9 fold higher, respectively, than those of control bees (Figs. 3 A and B). The difference between the expression levels in the 24h treated (n=42) and control (n=42) bees was significant (ABA, P=0.044 and DEF, P=0.029). The differences in mRNA levels of both ABA and DEF at the other time points, 3, 6, 12 and 48 h post-inoculation, were not significant (n≥30, P>0.05). HYM transcript in adult bees did not change significantly as a result of KSBV infection at any time points (n≥30, P>0.05); however, there was a trend toward a higher expression level between 6h and 48h after infection, when compared to controls (Fig. 1C).
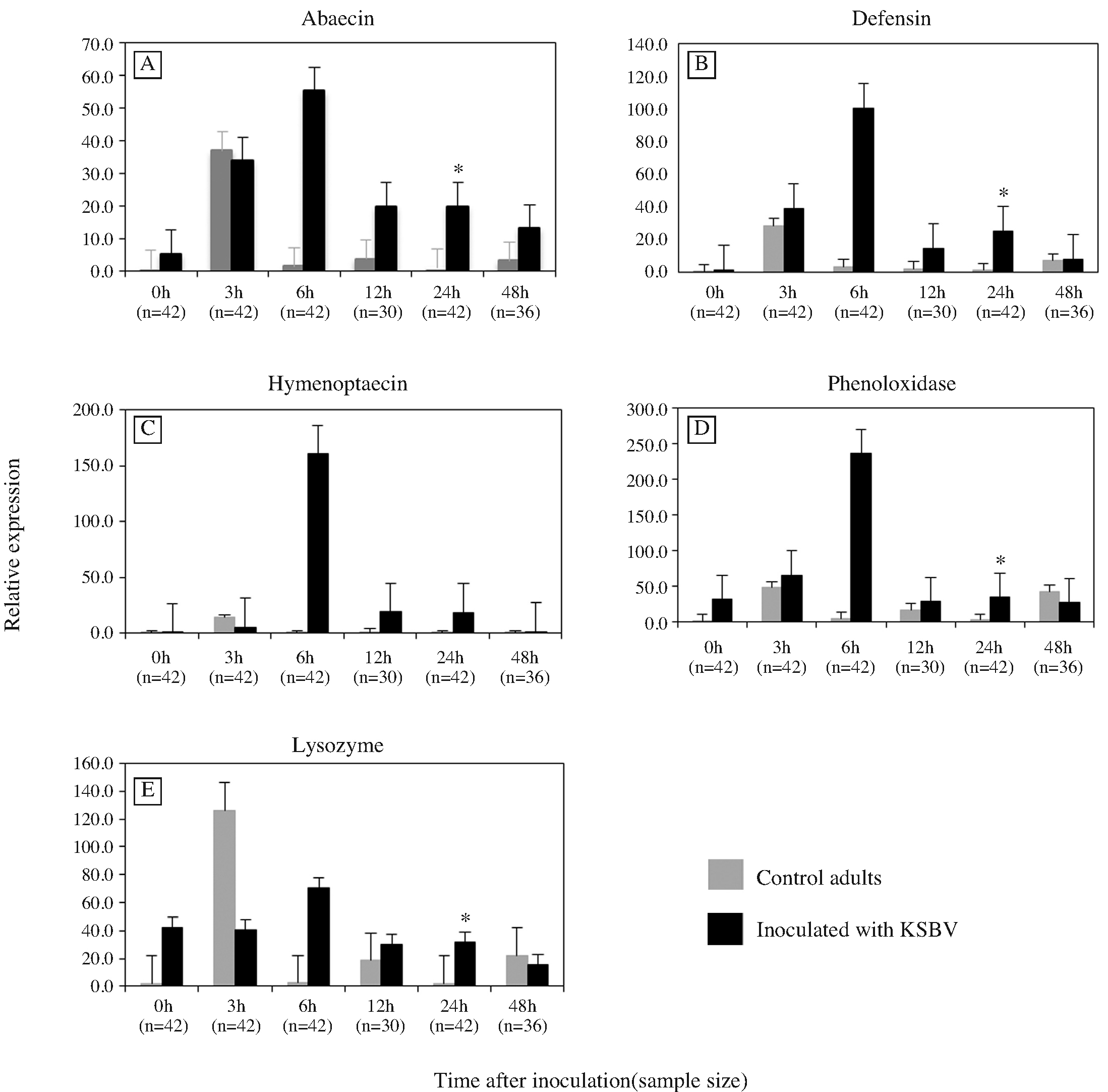
Expression levels of three antimicrobial peptide genes (A) abaecin (B) defensin and (C) hymenoptaecin, and two immune-related enzyme genes (D) phenoloxidase and (E) lysozyme, in control adult bees and adult bees exposed to Korean sacbrood virus (KSBV) measured by qRT-PCR at 0, 3, 6, 12, 24 and 48 h post infection. Gene transcript concentrations were normalized to the level of β-actin transcript in each sample. Positive standard errors are shown above each column. Asterisks indicate statistical significance by ANOVA (P<0.05).
In the bee larvae, the expression changes of the three AMPs genes in response to KSBV infection had most downregulated at experimental time points except ABA at 24 h post infection. However, there were no significant expression changes detected between KSBV-infected and control larvae although a down-regulation was observed in ABA level (3, 6, 12 h), DEF and HYM levels at 3, 6, 12 and 24 h after exposure to KSBV when compared to the controls in four trials (Fig. 2A, B and C).
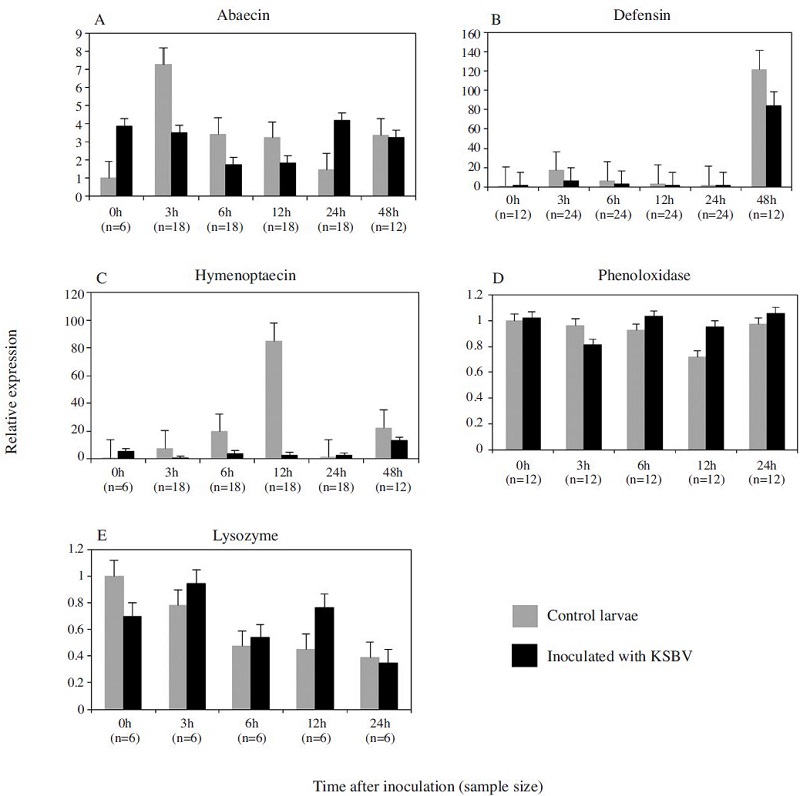
Expression levels of three antimicrobial peptide genes (A) abaecin (B) defensin and (C) hymenoptaecin, and two immune-related enzyme genes (D) phenoloxidase and (E) lysozyme, in control larvae and larvae exposed to Korean sacbrood virus (KSBV) measured by qRT-PCR at 0, 3, 6, 12, 24 and 48 h post infection. Gene transcript concentrations were normalized to the level of β-actin transcript in each sample. Positive standard errors are shown above each column.
Expression levels of two immune-related enzyme genes in honeybees upon KSBV infection
The patterns of PO and LYS gene expression in adult bees infected with KSBV had a similar trend (Fig. 1D and Fig. E) in seven independent experiments. The mRNA levels of PO and LYS were 18.08 and 16.03 fold higher, respectively, 24 h post-inoculation when compared with control adults (Fig. 3D and E). These changes were significantly different between the infected (n=42) and control (n=42) bees (PO, P=0.019 and LYS, P=0.026); however, both PO and LYS were down regulated at 48 h after challenge with KSBV. In the two independent experiments with bee larvae, the mRNA expression levels of PO and LYS were slightly higher (< 2-fold) at 6 h and 12 h following infection with KSBV when compared with controls. However, the differences were not significant. In larvae, PO and LYS mRNA levels were not measured beyond 24 h exposure to KSBV (Figs. 2D and E).
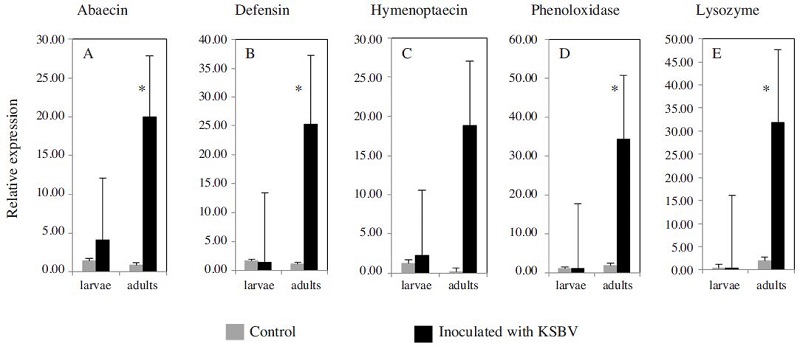
Expression levels of three antimicrobial peptide genes (A) abaecin (B) defensin and (C) hymenoptaecin, and two immune-related enzyme genes (D) phenoloxidase and (E) lysozyme in control larvae and adult bees, and in larvae and adult bees exposed to Korean sacbrood virus (KSBV) for 24 h. Positive standard errors are shown above each column. Asterisks indicate statistical significance by ANOVA (P<0.05).
DISCUSSION
To date, studies of the immune response in A. mellifera have been mainly in relation to bacterial infection (Gatchenberger et al., 2012; Laughton et al., 2011; Randolt et al., 2008; Evans et al., 2004). There are some reports concerned with the immune response of A. mellifera to infection with mites (Garrido et al., 2013; Zhang et al., 2010; Yang et al., 2005) and protozoa (Chaimanee et al., 2012; Antunez et al., 2009). However, research in the field of honeybee virology has been limited by the lack of understanding of the mechanisms behind the defenses observed in bees. The RNAi mechanism is present in most organisms as a way of managing gene regulation and antiviral response (Hannon, 2002). Honeybees have been observed to invoke RNAi in response to Israeli acute paralysis virus (IAPV), another picornavirus (Hunter, 2010). Recently, the immune response of bees to acute bee paralysis virus (ABPV) and Chinese Sacbrood Virus (CSBV) have been investigated (Azzami et al., 2012; Zhang et al., 2013). Like SBV and other closely related viruses, KSBV, which causes death in both larvae and adult bees, is one of the most damaging infections that currently threaten Korean honeybee colonies. The primary aim of the present study was to assess the immune adaptability of A. mellifera in the face of KSBV infection by monitoring the gene expression of three small AMPs (ABA, DEF and HYM) and two enzymes with immune activity (PO and LYS). To our knowledge, this is the first study to evaluate the immune response of honeybees infected by KSBV.
We found that the three AMP genes were upregulated in adult bees infected with KSBV when compared to control adults, but only ABA and DEF levels were significantly different. In addition, the trend of upregulation was observed only in the adult worker bees and not the larvae, showing that there is clearly a difference in AMP gene expression between A. mellifera larvae and adults infected with KSBV. Zhang et al., (2013) analyzed 32 immunerelated genes in A. cerana challenged by CSBV. Among 22 genes which statistically changed, 11 genes related to RNAi pathway were up-regulated and other 11 genes related to Toll, lmd, and Jak-Stat pathway were downregulated. While a previous study by Azzami et al. (2012) investigated of these three proteins at translational level in response to ABPV, it did not present evidence for detectable AMP production in bee larvae and adults at any time post-infection. In fact, AMPs are components of innate immunity, forming the first-line of defense used by many organisms against invading pathogens (Jenssen et al., 2006). AMPs-induced responses require specific pathogen recognition to stimulate their production. ABA is known to be effective against Gram-positive bacteria through permeabilizing the outer bacterial membrane (Casteels et al., 1990); DEF in honeybee is classified into two sub families: antibacterial and antifungal. Antibacterial DEF is efficacious against Grampositive bacteria (Casteels et al., 1994); meanwhile, antifungal defensins are predominantly effective against filamentous fungi (Bulet and Stocklin 2005). And of the known honeybee-specific AMPs, HYM plays a central role because it affects a wide variety of Gram-positive and Gram-negative bacteria by sequential permeabilization of the bacterial outer and inner membrane (Casteels et al., 1993). Upregulation of AMP production in bees is an indication of an active humoral immune response against bacterial invasion (Evans, 2004; Randolt et al., 2008). Viruses lack the stimulatory cellular membrane structure of bacteria, and viral capsid diversity provides no known conserved target for AMPs, yet we found AMP upregulation in response to KSBV. This finding suggests that if in A. mellifera these AMPs are being induced to combat KSBV it is most likely as part of an indirect response, for example to promote the expression of other AMPs. Otherwise, these AMP genes were induced by either different experimental conditions or own properties of KSBV. Therefore, the different expression of AMPs is not important for the immune response of A. mellifera to KSBV.
We also used qRT-PCR to determine the transcript abundance of the immunerelated enzymes, PO and LYS. PO is a vital enzyme essential for a number of processes such as sclerotization, wound healing, nodulation and phagocytosis stimulation. PO-catalyzed melanization of pathogens is a key element of invertebrate defenses (Cerenius et al., 2008), and may compensate for the absence of specific cellular defense mechanisms. In the present study, as shown in Fig. 1D and 2D, PO was upregulated at almost all time points after challenge with KSBV. Compared to control adult bees, significant upregulation (18.08-fold) occurred in KSBV-infected adult bees after 24 h exposure (3D). Although expression was upregulated in larvae infected with KSBV, we did not see a significant change (more than 1-fold). According to experiments reported by Evans and colleagues (2006b), expression of the PO gene is upregulated in natural bacterial infection but not with artificial challenge. We need to test more than the samples in order to address this difference. On the other hand, in our experiments PO expression was very low in both control and infected larvae but not in adults (2D and 1D), concordant with the findings of others (Lourenco et al., 2005; Laughton et al., 2011). According to Lourenco and colleagues (2005), the amount of proPO transcript in larvae and the pupae of worker bees start low and increases with age. Others report that as well as the level of proPO transcript rising, PO activity increases steadily with age in adult workers and queens (Wilson- Rich et al., 2008; Schmid et al., 2008; Laughton et al., 2011). In addition, Wilson et al. (2001) showed that high levels of midgut PO correlated with increased resi-stance to a baculovirus in lepidoptera. This is in line with the work of Washbuen et al. (1996, 2000) which indicates that the spread of baculovirus in insects is blocked by the aggregation of hemocytes forming melanotic capsules around infected cells, and that melanin is also produced during this process, suggesting a role for PO. Taken together, these findings imply that PO most likely plays a significant role in the antiviral responses of adult honeybees.
The synthesis of LYS was significantly increased not only in adult bees but also in newly emerged worker bees upon KSBV challenge (Fig. 1E). Lysozyme is muramidase that hydrolyse the β-1, 4-glycosidic linkage in the N-acetyl glucosamine and N-acetyl muramic acid residues in the peptidoglycan layer of the bacterial cell and cause their lysis, thereby playing an important role in antibacterial defense (Bachali et al., 2002). Expression of the LYS gene is strongly induced by bacteria (Hultmark, 1996; Gillespie et al., 1997). Interestingly, we detected upregulation of LYS in both larvae and adult worker bees when infected with KSBV, as shown in Fig. 1E and 2E. However, the level of change in adults was much higher than that in larvae. We also could not explain the upregulation of LYS in adult bees infected with KSBV because its difference was significant (P=0.026). Hence, as in the case of AMPs, we think it likely that LYS is directly involved in the honeybee defense response to KSBV because it is known to promote the expression of AMPs (Imler and Bulet, 2005).
In general, of the genes examined in this study most showed upregulated expression in adult bees infected with KSBV while only a few changes in expression occurred in infected larvae. Although the function or role of these genes induced by KSBV infection in both larvae and adult worker bees are not all easily explained, these results provide information about the immune response of A. mellifera to KSBV.
Acknowledgments
This study was supported by Animal and Plant Quarantine Agency and Bio-industry Technology Development Program (Nr. 312027-3), Ministry of Agriculture, Food and Rural Affairs.
LITERATURE CITED
-
Allen, M., Ball, B., (1996), The incidence and world distribution of honey bee viruses, Bee World, 77, p141-162.
[https://doi.org/10.1080/0005772X.1996.11099306]

-
Antunez, K., Martin-Hernandez, R., Prieto, L., Meana, A., Zunino, P., Higes, M., (2009), Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (microsporidia), Enviromental Microbiology, 11, p2284-2290.
[https://doi.org/10.1111/j.1462-2920.2009.01953.x]

- Aupinel, P., Fortini, D., Dufour, H., Tasei, J. N., Michaud, B., Odoux, J.F., Pham-Delegue, M.H., (2005), Improvement of artificial feeding in a standard in vitro method for rearing Apis mellifera larvae, Bulletin of Insectology, 58, p107-111.
-
Azzami, K., Ritter, W., Tautz, J., Beier, H., (2012), Infection of honey bees with acute bee paralysis virus does not trigger humoral or cellular immune responses, Arch. Virol, 157, p689-702.
[https://doi.org/10.1007/s00705-012-1223-0]

-
Bachali, S., Jager, M., Hassanin, A., Schoentgen, F., Jolles, P., Fiala-Medioni, A., Deutsch, J. S., (2002), Phylogenetic analysis of invertebrate lysozymes and the evolution of lysozyme function, J. Mol. Evol, 54, p652-664.
[https://doi.org/10.1007/s00239-001-0061-6]

- Bailey, L., Ball, B.V., (1991), Honey Bee Pathology, Academic Press, London.
- Ball, B.V., (1999), Sacbrood, In: Colin, ME (ed.), Ball, BV (ed.), Kilani, M. (eds), Bee Disease Diagnosis Zaragoza, CIHEAM, p91-97.
-
Bulet, P., Stocklin, R., (2005), Insect antimicrobial peptides: structures, properties and gene regulation, Protein and Peptide Letters, 12, p3-11.
[https://doi.org/10.2174/0929866053406011]

- Casteels, P., Ampe, C., Jacobs, F., Tempst, P., (1993), Functional and chemical of Hymenoptaecin, an antibacterial polypeptide that is infection-inducible in the honeybee (Apis mellifera), J. Biol. Chen, 268, p7044-7054.
-
Casteels, P., Ampe, C., Riviere, L., Van Damme, J., Elicone, C., Fleming, M., Jacobs, F., Tempst, P., (1990), Isolation and characterization of abaecin, a major antibacterial response peptide in the honeybee (Apis mellifera), Eur. J, Biochem.187, p381-386.
[https://doi.org/10.1111/j.1432-1033.1990.tb15315.x]

- Casteels-Jonsson, K., Zhang, W., Capaci, T., Casteels, P., Tempst, P., (1994), Acute transcriptional response of the honeybee peptide-antibiotics gene repertoire posttranslational conversion of the precursor structures, J. Biol. Chem, 269s, p28569-28575.
-
Cerenius, L., Lee, B.L., Soderhall, K., (2008), The proPOsystem: pros and cons for its role in invertebrate immunity, Trends Immunol, 29, p263-271.
[https://doi.org/10.1016/j.it.2008.02.009]

-
Chaimanee, V., Chantawannakul, P., Chen, Y., Evans, J.D., Pettis, J.S., (2012), Differential expression of immune genes of adult honey bee (Apis mellifera) after inoculated by Nosema ceranae, Journal of Insect Physiology, 58, p1090-1095.
[https://doi.org/10.1016/j.jinsphys.2012.04.016]

-
Chen, Y.P., ?Siede, R., (2007), Honey bee viruses, Adv. Virus Res, 70, p33-80.
[https://doi.org/10.1016/s0065-3527(07)70002-7]

-
Choe, S.E., Nguyen, T.T.D., Hyun, B.H., Noh, J.H., Lee, H.S., Lee, C.H., Kang, S.W., (2012), Genetic and phylogenetic analysis of South Korea sacbrood virus isolates from infected honey bees (Apis cerana), Veterinary Microbiology, 157, p32-40.
[https://doi.org/10.1016/j.vetmic.2011.12.007]

-
De Miranda, J.R., Bailey, L., Ball, B.V., Blanchard, P., Budge, G., Chejanovsky, N., Chen, Y-P., Gauthier, L., Genersch, E., De Graaf, D., Ribiere, M., Ryabov, E., De Smet, L., Van Der Steen, J. J. M., (2013), Standard methods for virus research in Apis mellifera, In V. Dietemann, J D. Ellis, P. Neumann (Eds), The COLOSS BEEBOOK, IIVolume : standard methods for Apis mellifera pest and pathogen research, Journal of Apicultural Research, 52(4), http://dx.doi.org/10.3896/IBRA.1.52.4.22.
[https://doi.org/10.3896/IBRA.1.52.4.22]

-
Decker, H., Jaenicke, E., (2004), Recent findings on phenoloxidase activity and antimicrobial activity of hemocyanins, Dev. Comp. Immunol, 28, p673-687.
[https://doi.org/10.1016/j.dci.2003.11.007]

-
Evan, J. D., (2004), Transcriptional immune responses by honey bee larvae during invasion by the bacterial pathogen, PaeniBacillus larvae, J. Invertebr. Pathol, 85, p105-111.
[https://doi.org/10.1016/j.jip.2004.02.004]

-
Evans, J.D., and Pettis, J.S., (2005), Colony-level impacts of immune responsiveness in honey bees, Apis mellifera, Evolution, 59, p2270-2274.
[https://doi.org/10.1111/j.0014-3820.2005.tb00935.x]

-
Evans, J.D., (2006a), Bee path: an ordered quantitative-PCR array for exploring honey bee immunity and disease, J. Invertebr. Pathol, 93, p135-139.
[https://doi.org/10.1016/j.jip.2006.04.004]

-
Evans, J.D., Aronstein, K., Chen, P.Y., Hetru, C., Imler, J.L., Jiang, H., Kanost, M., Thompson, G.J., Zou, Z., Hulmark, D., (2006b), Immune pathways and defence mechanisms in honey bees Apis mellifera, Insect Molecular Biology, 15, p645-656.
[https://doi.org/10.1111/j.1365-2583.2006.00682.x]

-
Garrido, P.M., Antunez, K., Martin, M., Porrini, M.P., Zunino, P., Eguaras, M.J., (2013), Immune-related gene expression in nurse honey bees (Apis mellifera) exposed to synthetic acaricides, J. Insect Physiol, 59, p113-119.
[https://doi.org/10.1016/j.jinsphys.2012.10.019]

-
Gatschenberger, H., Gimple, O., Tautz, J., Beier, H., (2012), Honey bee drones maintain humoral immune competence throughout all life stages in the absence of vitellogenin production, J. Exp. Biol, 215, p1313-1322.
[https://doi.org/10.1242/jeb.065276]

-
Gillespie, J. P., Kanost, M. R., and Trenczek, T., (1997), Biological mediators of insect immunity, Annu. Rev. Entomol, 42, p611-643.
[https://doi.org/10.1146/annurev.ento.42.1.611]

-
Hannon, G. J., (2002), RNA interference, Nature, 418, p24-26.
[https://doi.org/10.1038/418244a]

-
Hoffmann, J. A., (2003), The immune response of Drosophila, Nature, 426, p33-38.
[https://doi.org/10.1038/nature02021]

- Hultmark, D., (1996), Insect lysozymes, in: Jolles, P. (Ed), Lysozymes: Model Enzymes in Biochemistry and Biology, Birkhauser Verlag, Basel, p87-102.
-
Hunter, W., (2010), Medium for development of bee cell cultures (Apis mellifera: Hymenoptera: Apidae), In Vitro Cellular & Developmental Biology-Animal, 46, p83-86.
[https://doi.org/10.1007/s11626-009-9246-x]

-
Imler, J.L., and Bulet, P., (2005), Antimicrobial peptides in Drosophila: structures, activities and gene regulation, in: Kabelitz, D., and Schorder, J.M. (Eds), Mechanism of Epithelial Defense, 86, Basel, Karger, p1-21.
[https://doi.org/10.1159/000086648]

-
Jenssen, H., Hamill, P., Hancock, R.E., (2006), Peptide antimicrobial agents, Clin. Microbiol. Rev, 19, p491-511.
[https://doi.org/10.1128/CMR.00056-05]

-
Laughton, A.M., Boots, M., Siva-Jothy, M.T., (2011), The ontogeny of immunity in the honey bee, Apis mellifera L. following an immune challenge, J. Insect Physiol, 57, p1023-1032.
[https://doi.org/10.1016/j.jinsphys.2011.04.020]

-
Lourenco, A.P., Zufelato, M.S., Bitondi, M.M.G., Simoes, Z.L.P., (2005), Molecular characterization of a cDNA encoding prophenoloxidase and its expression in Apis mellifera, Insect Biochem. Mol. Biol, 35, p541-552.
[https://doi.org/10.1016/j.ibmb.2005.01.013]

-
Peng, Y-S.C., Mussen, E., Fong, A., Montague, M.A., Tyler, T., (1992), Effects of chlortetracycline of honey bee worker larvae reared in vitro, J. Invertebr. Pathol, 60, p127-133.
[https://doi.org/10.1016/0022-2011(92)90085-I]

-
Randolt, K., Gimple, O., Geissendorfer, J., Reinders, J., Prusko, C., Mueller, M.J., Albert, S., Tautz, J., Beier, H., (2008), Immune-related proteins induced in the hemolymph after aseptic and septic injuries differ in honey bee worker larvae and adults, Arch. Insect Biochem. Physiol, 69, p155-167.
[https://doi.org/10.1002/arch.20269]

-
Schmid, M.R., Brockmann, A., Pirk, C.W.W., Stanley, D.W., and Tautz, J., (2008), Adult honeybees (Apis mellifera L.) abandon hemocytic, but not phenoloxidase based immunity, J. Insect Physiol, 54, p439-444.
[https://doi.org/10.1016/j.jinsphys.2007.11.002]

-
Washburn, J.O., Haas-Stapleton, E.J., Tan, F.F., Beckage, N.E., Volkman, L.E., (2000), Co-infection of Manduca sexta larvae with polydnavirus from Cotesia congregata increases susceptibility to fatal infection by Autographa californica M Nucleopolyhedrovirus, J. Insect Physiol, 46, p179-190.
[https://doi.org/10.1016/S0022-1910(99)00115-8]

-
Washburn, J.O., Kirkpatrick, B.A., Volkman, L.E., (1996), Insect protection against viruses, Nature, 383, p767.
[https://doi.org/10.1038/383767a0]

-
Wilson, K., Cotter, S.C., Reeson, A.F., Pell, J.K., (2001), Melanism and disease resistance in insects, Ecology Letters, 4, p637-649.
[https://doi.org/10.1046/j.1461-0248.2001.00279.x]

-
Wilson-Rich, N., Dres, S.T., Starks, P.T., (2008), The ontogeny of immunity: Development of innate immune strength in the honey bee (Apis mellifera), J. Insect Physiol, 54, p1392-1399.
[https://doi.org/10.1016/j.jinsphys.2008.07.016]

-
Yang, X., Cox-Foster, D.L., (2005), Impact of an ectoparasite on the immunity and pathology of an invertebrate: evidence for host immunosuppression and viral amplification, PNAS, 21, p7470-7475.
[https://doi.org/10.1073/pnas.0501860102]

-
Zhang, Y., Huang, X., Xu, Z.F., Han, R.C., Chen, J.H., (2013), Differential Gene Transcription in Honeybee (Apis cerana) Larvae Challenged by Chinese Sacbrood Virus (CSBV), Sociobiology, 60, p413-420.
[https://doi.org/10.13102/sociobiology.v60i4.413-420]

-
Zhang, Y., Liu, X., Zhang, W., Han, R., (2010), Differential gene expression of the honey bees Apis mellifera and A. cerana induced by Varroa destructor infection, J. Insect Physiol, 56, p1207-1218.
[https://doi.org/10.1016/j.jinsphys.2010.03.019]

