Virome Profile Uncovers the First Identification of Lake Sinai Virus in the Asian Honey Bee (Apis cerana; Hymenoptera: Apidae) in South Korea
Abstract
The Asian honey bee (Apis cerana) is a keystone pollinator in East and South Asia, crucial to regional biodiversity and agricultural productivity. Despite its ecological significance, A. cerana has been severely impacted by viral pathogens, particularly sacbrood virus (SBV), which has decimated colonies in South Korea. Here, we report the first detection of Lake Sinai virus (LSV) in A. cerana populations in South Korea, marking a substantial addition to our understanding of LSV host range and interspecies transmission dynamics. Using high-throughput sequencing (HTS)-based virome analysis, we identified three distinct LSV species-LSV2, LSV3, and LSV4-alongside eight other honey bee-associated viruses. This study provides evidence of potential LSV transmission from A. mellifera, where it was previously detected in South Korea, expanding the virome landscape of A. cerana. These findings highlight the urgent need for ongoing virological surveillance in A. cerana populations to track emerging viral threats. This study underpins critical strategies for enhancing honey bee health, conservation, and disease management to secure the pollination services essential for ecosystem stability.
Keywords:
Apis cerana, Virome analysis, High-throughput sequencing, Lake Sinai virus, Honey bee virus, Viral spilloverINTRODUCTION
The Asian honey bee, Apis cerana, holds an indispensable ecological role as a primary pollinator in diverse environments across East and South Asia (Egelie et al., 2015). Noted for its adaptability and resilience in varied climatic conditions, A. cerana supports both agricultural and natural ecosystems. Unlike the Western honey bee (A. mellifera), which has been extensively studied and managed in apiculture, A. cerana demonstrates unique immunity characteristics and ecological adaptability, enabling survival in challenging environments such as mountainous and subtropical regions (Chen et al., 2018). Despite producing less honey, A. cerana contributes significantly to biodiversity, making its conservation essential for ecological and agricultural stability (Koetz, 2013; Zhang et al., 2019; Donkersley et al., 2021; Nannan et al., 2022; Katuwal et al., 2023).
In South Korea, A. cerana faces a high prevalence of viral infections, with sacbrood virus (SBV) and black queen cell virus (BQCV) causing substantial colony losses (Choe et al., 2012). The impact of SBV is particularly devastating, with mortality rates reaching up to 90%, prompting initiatives to develop SBV-resistant strains. However, other viruses remain a growing threat, underscoring the importance of broad-spectrum monitoring and virome analysis to understand the health dynamics within A. cerana populations (Vung et al., 2017).
High-throughput sequencing (HTS) revolutionized genomics and virus detection, offering unparalleled sensitivity and precision over traditional sequencing methods (Pérez-Losada et al., 2020; Fitzpatrick et al., 2021). This technology allows researchers to generate vast datasets rapidly, facilitating early detection of novel viruses and swift responses to emerging epidemics. As HTS technology has advanced, it has become a cornerstone in viral research worldwide, enabling the efficient identification of a diverse array of viruses and their variants (Roberts et al., 2018; Massart et al., 2019; Bester et al., 2021).
Virome analysis, an application of HTS, plays a pivotal role in exploring viral diversity within specific ecosystems or host populations, uncovering interactions between viruses and their environment (Wolf et al., 2018; Li et al., 2022). In pollinators like honey bees, virome studies are essential for promoting ecological stability and sustaining biodiversity (Fetters and Ashman, 2023; Van Herzele et al., 2024). By revealing previously unknown viruses and tracing disease transmission pathways, virome analysis enhances our understanding of viral dynamics and interactions (Villamor et al., 2019; Kwon et al., 2023, 2024). Additionally, this approach aids in assessing the impact of specific viruses on honey bee health, informing strategies to prevent potential disease outbreaks (Kwon et al., 2023). Ultimately, virome analysis not only enriches our knowledge of viral ecology but also supports the development of targeted interventions to protect these vital pollinators (Li et al., 2023; Sbardellati and Vannette, 2024).
The alarming decline in honey bee populations has raised global concerns due to its implications for ecosystem stability and agricultural productivity. A. cerana, an essential pollinator species in Asia, plays a critical role in these systems, yet research on this species remains limited. Recent discoveries of various viruses affecting A. cerana in China underscore the need for further studies to evaluate the impact of these infections on populations in South Korea (Li et al., 2023). This study seeks to systematically identify and analyze the virome of A. cerana in South Korea, employing advanced sequencing technologies to address health challenges and inform strategies to mitigate the adverse effects of viral infections on these populations.
MATERIALS AND METHODS
1. Sample collection and preparation for sequencing
Honey bee adults were collected from five randomly selected Apis cerana colonies located in Busan, South Korea. The bees were stored in 50 mL conical tubes containing 70% ethanol (EtOH) and transported to the laboratory under refrigerated conditions. Upon arrival, the samples were stored at 4℃. Two honey bees were randomly selected from each conical tube, and the ethanol residue was removed by gently blotting them with tissue paper. The whole bodies of honey bees were placed in sample bags (1523W, 3M, Maplewood, MN, USA) containing 10 mL of PCR-grade water (Tech & Innovation, Chuncheon, South Korea) and homogenized at 10 m/s for 40 seconds using a stomacher blender (LC-08, LabTech, Jeonju, South Korea). Any uneven particles remaining after homogenization were further smoothed out using a roller. One milliliter of the homogenate, excluding exoskeletal fragments, was transferred to 1.5 mL tubes. Total RNA extraction was conducted with TRIzol® (Invitrogen, Carlsbad, CA, USA), and the purity and concentration were assessed using a NanoPhotometer® NP80 (IMPLEN GmbH, Munich, Germany). RNA was quantified to a concentration of 300 ng/μL for library preparation, and remaining samples were stored at -80℃ for future PCR analyses.
2. High-throughput sequencing (HTS) and virome analysis
Libraries were prepared using the TruSeq Stranded Total RNA with Ribo-Zero H/M/R Gold kit (Illumina, San Diego, CA, USA) and assessed for quality using a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). HTS was performed on an Illumina NovaSeq 6000 system. Raw FASTQ files were processed using CLC Genomics Workbench (ver. 24.0.1, QIAGEN, Hilden, Germany) for virome analysis. Barcode sequences and low-quality reads were trimmed, and host sequences were removed using the A. cerana genome from the NCBI GenBank database to retain unmapped reads. De novo assembly of these reads generated contigs, which were analyzed using tBLASTx against viral sequences in the NCBI GenBank database. Contigs with significant E-values (0 to xe-10) were extracted and subjected to BLASTn analysis with a database of non-human host viruses from the NCBI virus resource. The viruses identified were subsequently confirmed by mapping the unmapped reads to the viral sequence database.
3. Virus verification by RT-PCR
For RT-PCR verification, virus-specific primers were designed using Primer3 (https://primer3.ut.ee/, Table 1) if existing primers were unavailable. RT-PCR reactions were conducted with SuPrimeScript RT-PCR premix (2×) from GENETBIO (Daejeon, South Korea). The presence of viral sequences was confirmed by gel electrophoresis on a 1% agarose gel.
4. Phylogenetic classification of viral sequences
Viral genome sequences from A. cerana were aligned to reference sequences in the NCBI GenBank database using MAFFT (Katoh et al., 2002). IQ-TREE2 software was employed to select the optimal model, and phylogenetic trees were constructed with 1000 bootstrap replicates (Minh et al., 2020). Visualization of phylogenetic relationships was performed with iTOL, elucidating the evolutionary relationships among detected viruses (Letunic and Bork, 2024).
RESULTS
1. High-throughput sequencing (HTS) and virome analysis
HTS generated a total of 5.64 GB of data, yielding 59,959,882 high-quality reads, with a Phred quality score over 20 (Q20) in 98.2% of bases and over 30 (Q30) in 94.8% of bases. After quality trimming, 59,943,883 reads remained, of which 34,116,664 were host-related, while 116,984 reads were identified as viral sequences (Fig. 1). Virome analysis identified eight distinct viruses: Apis mellifera-associated partiti-like virus 1 (AmPLV1), black queen cell virus (BQCV), deformed wing virus (DWV), Hubei partiti-like virus 34 (HPLV34), Lake Sinai virus (LSV) 2, 3, and 4, and sacbrood virus (SBV). Among these, DWV had the highest read count, while complete genome sequences were obtained for AmPLV1 and SBV.
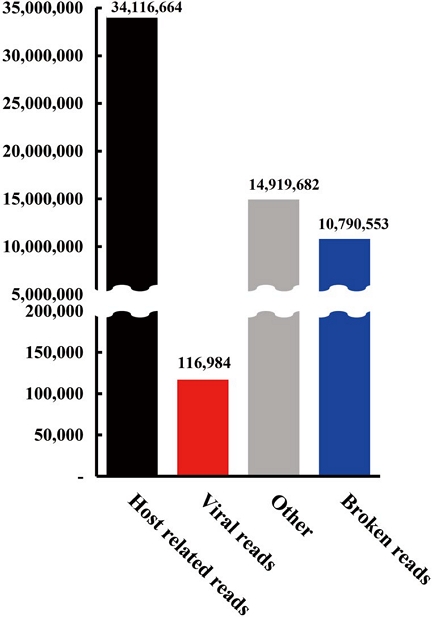
Distribution of reads across categories from virome analysis. The bar chart illustrates the distribution of reads among different categories in the virome dataset. The red bar indicates viral reads identified using reference viral genome sequences from the NCBI GenBank database, emphasizing the proportion of viral content in the overall dataset.
2. Genetic characterization of Lake Sinai viruses (LSVs)
This study reports the first detection of LSV in A. cerana in South Korea. Virome analysis identified three LSV species (LSV2, LSV3, and LSV4) with read counts of 175, 204, and 110, respectively, suggesting low abundance. Partial sequences were obtained for all three species (Table 2). Using reference templates MZ821876, OR496475, and OP972894, the assemblies achieved 84% coverage for LSV2, 88% for LSV3, and 70% for LSV4, with open reading frames (ORFs) for ORF1, RNA-dependent RNA polymerase (RdRp), and capsid protein confirmed (Fig. 2). BLASTn analysis revealed that LSV2 was 94.62% similar to MT732482 from China and had over 93% similarity with South Korean sequences. LSV3 showed a 97.16% identity with South Korean sequence OR496477 and over 96% similarity with Chinese sequences but only 83-85% similarity with European variants. LSV4 had 98.21% identity with South Korean sequence OR496491 and above 95% similarity with Chinese sequences (Table 2).
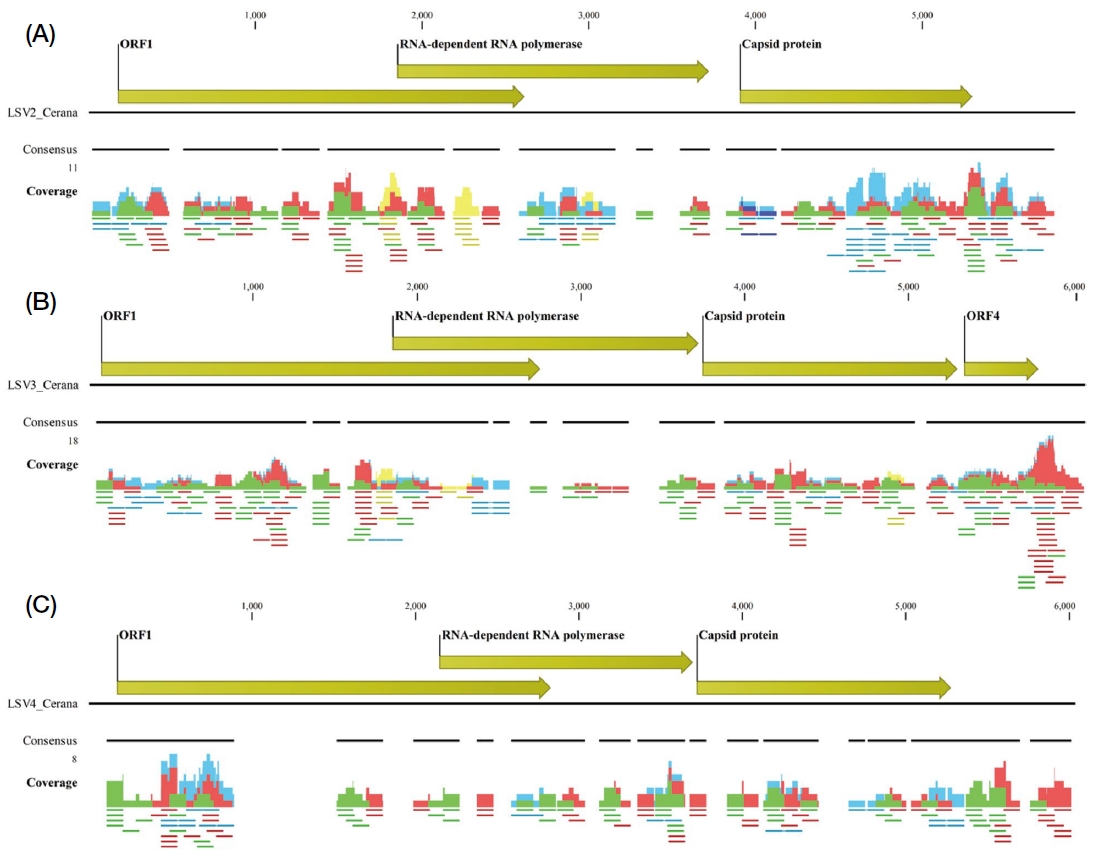
Genome organization and sequencing coverage of LSV2, LSV3, and LSV4 in Apis cerana. The genome structures of three A. cerana-associated viruses (LSV2, LSV3, and LSV4) are displayed with annotated open reading frames (ORFs) for ORF1, RNA-dependent RNA polymerase, capsid protein, and ORF4. Coverage plots show sequencing depth with colors representing read types: paired reads (blue), forward unpaired reads (green), reverse unpaired reads (red), and non-specific matches (yellow). Consensus regions are marked with black bars below each coverage plot.
3. Genetic characterization of other honey bee viruses
Beyond LSV, additional viruses including AmPLV1, BQCV, DWV-A, HPLV34, and SBV were identified. Complete sequences were achieved for AmPLV1 and SBV, while others showed over 90% sequence coverage. A new sequence of AmPLV1 was identified with 96.82% similarity to OP972921. BQCV was most similar to OR496411 (99.06% similarity). DWV-A showed 97.09% similarity with OP972875, while type B was only 85% homologous. HPLV34, detected in A. cerana for the first time, shared 99.87% similarity with OR496448. SBV displayed high homology of 98.55% with PQ376985 (Table 2).
4. PCR validation and correlation with virome analysis
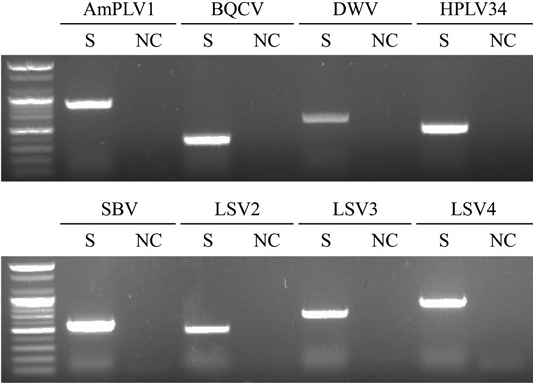
PCR targeting eight viruses confirmed the presence of each virus, with amplification products verified through 1% agarose gel electrophoresis (Fig. 3). Amplified bands reflected relative abundance, with stronger bands for viruses with higher read counts, supporting a correlation between PCR and HTS results.
5. Comparative phylogenetic and BLAST results
Phylogenetic analysis of seven viruses revealed differences between BLAST and phylogenetic similarities. For instance, while BLAST showed close similarity between LSV2 and MT732482, phylogenetically, LSV2 was more aligned with sequences from South Korea and China, diverging from MT732482 (Fig. 4A). Similarly, LSV3 was closest to OR496477 by BLAST but phylogenetically nearer to OR496480 (Fig. 4B). LSV4 showed a closer phylogenetic relationship to OP972894 than to its BLAST-similar sequence, OR496491 (Fig. 4C). For BQCV, both methods aligned, showing close clustering with OR496 411 and OP972872 (Fig. 5A). Similarly, DWV, HPLV34, and SBV showed congruent clustering with their nearest sequences (Figs. 5B-D).
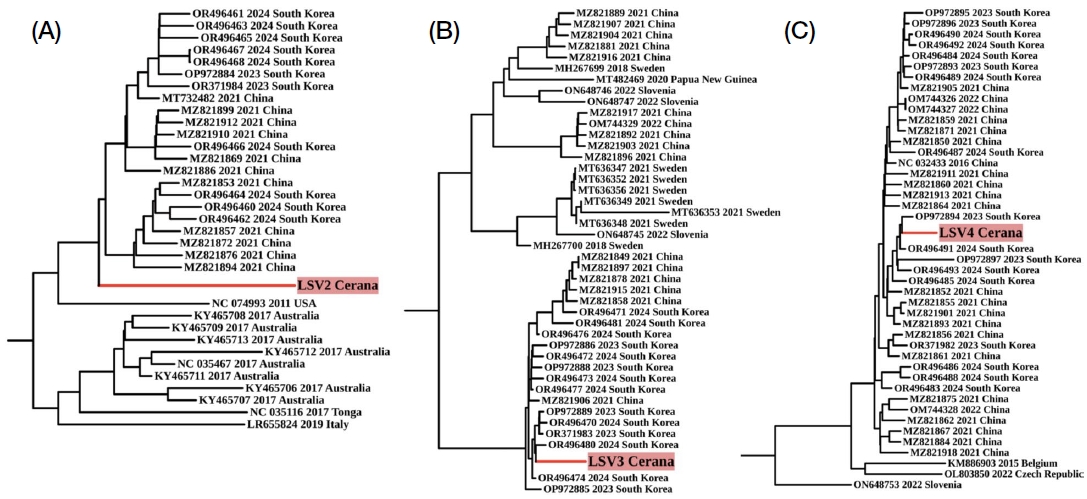
Maximum likelihood phylogenetic trees of Lake Sinai viruses (LSVs) constructed using IQ-TREE2 with 1,000 bootstrap replicates. Models were selected based on the Bayesian Information Criterion (BIC): (A) LSV2 using the TN+F+I+G4 model, (B) LSV3 using the TIM3+F+I+G4 model, and (C) LSV4 using the TN+F+I+G4 model. Bootstrap support values at key nodes demonstrate high confidence in the tree topology. Viral sequences identified in this study are highlighted in red.
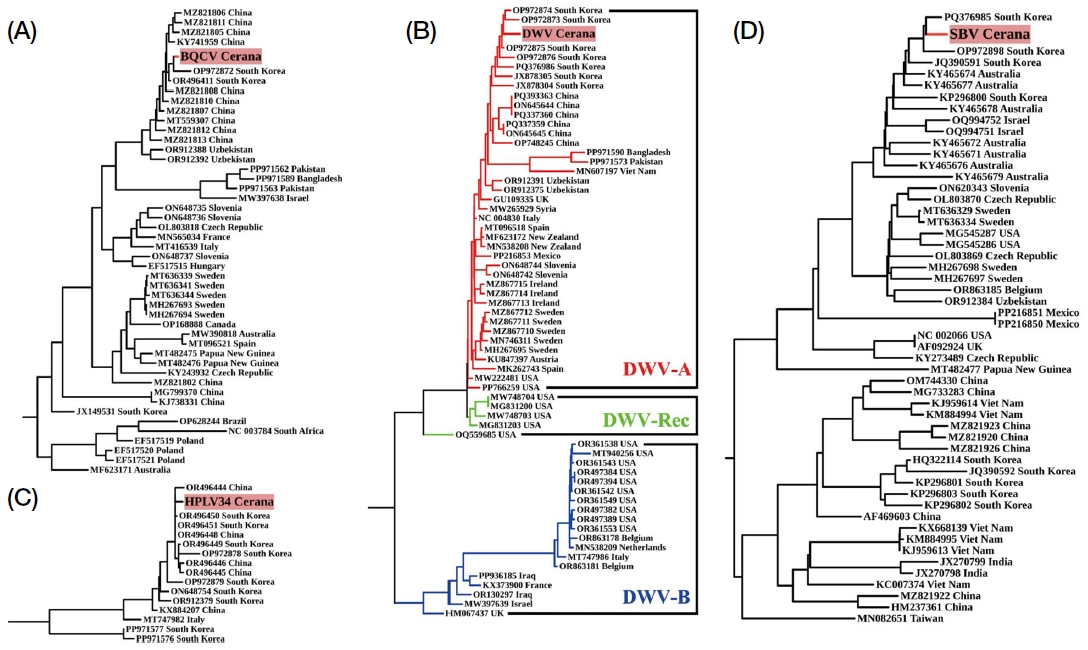
Maximum likelihood phylogenetic trees of honey bee viruses constructed using IQ-TREE2 with 1,000 bootstrap replicates. Models were chosen based on the Bayesian Information Criterion (BIC): (A) Black queen cell virus (BQCV) tree inferred with the GTR+F+I+G4 model, (B) Deformed wing virus (DWV) complex, including DWV-A, DWV-B, and DWV-Recombinant (DWV-Rec), with the TIM2+F+I+G4 model, (C) Hubei partiti-like virus 34 (HPLV34) tree with the HKY+F+I model, and (D) sacbrood virus (SBV) tree with the GTR+F+I+G4 model. Bootstrap values at key nodes indicate strong support for the inferred relationships. Viral sequences identified in this study are highlighted in red.
DISCUSSION
This study elucidates the viral landscape of A. cerana in South Korea, confirming multiple viruses, including SBV, BQCV, and DWV, which are known to negatively impact colony health. These viruses, particularly SBV, cause significant mortality in larvae and weaken adult bee immunity, threatening not only colony health but also pollination-dependent agriculture and ecological balance (Al Naggar and Paxton, 2020; Thu et al., 2020).
HTS-based virome analysis has proven essential in detecting both known and novel viruses, offering greater sensitivity than traditional PCR (Kwon et al., 2023). The detection of multiple LSV species, previously unidentified in A. cerana, expands our understanding of virus diversity and interspecies transmission, highlighting the role of virome analysis in advancing knowledge of viral evolution and distribution. Early pathogen detection through HTS-based virome analysis aids in developing surveillance and management strategies crucial for preserving bee health and agricultural ecosystems (Nikulin et al., 2024).
In this study, the presence of SBV, LSV3, and LSV4 was confirmed in A. cerana samples from South Korea, consistent with viruses previously identified in A. cerana populations in China (SAMN18146142 and SAMN181 46144). These findings highlight the shared viral landscape across Asian honey bee populations. Notably, this study marks the first detection of LSV2 in A. cerana, a virus previously found mainly in A. mellifera (Kwon et al., 2023). This new host detection suggests that factors such as environmental conditions, beekeeping practices, and genetic variation among bee populations may contribute to cross-species transmission of viral pathogens.
This discovery emphasizes the need to broaden virological research to include A. cerana alongside the more extensively studied A. mellifera. The identification of LSV2 in Korean A. cerana enriches our understanding of viral diversity and underscores the potential for cross-species viral transmission, underscoring the importance of expanding research focus on this species. Examining such occurrences can deepen our understanding of host-virus interactions and the evolutionary dynamics of viruses across various ecological and geographical settings. This research is essential for developing targeted strategies to conserve and manage bee populations globally, as they face diverse challenges across different environments.
Sinaivirus genus is a virus that has been identified not only in honey bees but also in various other insects, including weevils, hornet, and mantises (Shi et al., 2016; Toplak et al., 2020; François et al., 2021; Kitamura and Asai, 2022). However, comprehensive studies on its pathogenicity and associated symptoms in these hosts are still lacking. Further research is essential to better understand its impact and to develop strategies for mitigating potential future issues that may arise due to this virus.
While this study provides valuable insights into the viral profile of A. cerana in a specific region, it may not fully capture the broader geographical and environmental context. Future research should expand the sample range to comprehensively explore virus distribution and genetic diversity. Additionally, investigating the effects of mixed infections on bee physiology and behavior will be vital to understanding the combined threats to honey bee health. This integrated approach will offer a more complete view of the viral challenges facing bee populations worldwide, supporting the development of more effective management and conservation strategies.
Acknowledgments
This study was conducted as part of the “Glocal University Project Group in Andong National University-Gyeongbuk Provincial College” supported by the Ministry of Education and National Research Foundation of Korea.
AUTHOR CONTRIBUTIONS
E-JK and CJ conceived and designed the study. HO and CJ prepared the samples. MK and SY performed the experiments. E-JK and MK conducted bioinformatics and data analyses. MK and E-JK wrote the manuscript. All authors contributed to manuscript revision and have read and approved the submitted version.
References
-
Al Naggar, Y. and R. J. Paxton. 2020. Mode of transmission determines the virulence of black queen cell virus in adult honey bees, posing a future threat to bees and apiculture. Viruses 12(5): 535.
[https://doi.org/10.3390/v12050535]

-
Bester, R., G. Cook, J. H. Breytenbach, C. Steyn, R. De Bruyn and H. J. Maree. 2021. Towards the validation of high-throughput sequencing (HTS) for routine plant virus diagnostics: measurement of variation linked to HTS detection of citrus viruses and viroids. Virol. J. 18: 1-19.
[https://doi.org/10.1186/s12985-021-01523-1]

-
Chen, C., H. Wang, Z. Liu, X. Chen, J. Tang, F. Meng and W. Shi. 2018. Population genomics provide insights into the evolution and adaptation of the eastern honey bee (Apis cerana). Mol. Biol. Evol. 35(9): 2260-2271.
[https://doi.org/10.1093/molbev/msy130]

-
Choe, S. E., L. T. K. Nguyen, J. H. Noh, H. B. Koh, Y. H. Jean, C. H. Kweon and S. W. Kang. 2012. Prevalence and distribution of six bee viruses in Korean Apis cerana populations. J. Invertebr. Pathol. 109(3): 330-333.
[https://doi.org/10.1016/j.jip.2012.01.003]

-
Donkersley, P., L. Covell and T. Ota. 2021. Japanese honeybees (Apis cerana japonica Radoszkowski, 1877) may be resilient to land use change. Insects 12(8): 685.
[https://doi.org/10.3390/insects12080685]

- Egelie, A. A., A. N. Mortensen, J. L. Gillett-Kaufman and J. D. Ellis. 2015. Asian honey bee - Apis cerana. Entomology and Nematology Department, University of Florida. Retrieved 11.04 from https://entnemdept.ufl.edu/creatures/misc/bees/Apis_cerana.htm#top, .
-
Fetters, A. M. and T. L. Ashman. 2023. The pollen virome: a review of pollen-associated viruses and consequences for plants and their interactions with pollinators. Am. J. Bot. 110(6): e16144.
[https://doi.org/10.1002/ajb2.16144]

-
Fitzpatrick, A. H., A. Rupnik, H. O’Shea, F. Crispie, S. Keaveney and P. Cotter. 2021. High throughput sequencing for the detection and characterization of RNA viruses. Front. Microbiol. 12: 621719.
[https://doi.org/10.3389/fmicb.2021.621719]

-
François, S., A. Antoine-Lorquin, M. Kulikowski, M. Frayssinet, D. Filloux, E. Fernandez, P. Roumagnac, R. Froissart and M. Ogliastro. 2021. Characterisation of the viral community associated with the alfalfa weevil (Hypera postica) and its host plant, alfalfa (Medicago sativa). Viruses 13(5): 791.
[https://doi.org/10.3390/v13050791]

-
Katoh, K., K. Misawa, K. i. Kuma and T. Miyata. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14): 3059-3066.
[https://doi.org/10.1093/nar/gkf436]

-
Katuwal, D. R., A. Pokhrel and D. Khanal. 2023. Comparative Study of Apis cerena and Apis mellifera. J. Agric. For. Res. 2(3): 41-48.
[https://doi.org/10.2139/ssrn.4509061]

-
Kitamura, Y. and T. Asai. 2022. First detection of Lake Sinai virus in honeybees (Apis mellifera) and wild arthropods in Japan. J. Vet. Med. Sci. 84(3): 346-349.
[https://doi.org/10.1292/jvms.21-0466]

-
Koetz, A. H. 2013. Ecology, behaviour and control of Apis cerana with a focus on relevance to the Australian incursion. Insects 4(4): 558-592.
[https://doi.org/10.3390/insects4040558]

-
Kwon, M., C. Jung and E.-J. Kil. 2023. Metagenomic analysis of viromes in honey bee colonies (Apis mellifera; Hymenoptera: Apidae) after mass disappearance in Korea. Front Cell Infect Microbiol, 13, 27.
[https://doi.org/10.3389/fcimb.2023.1124596]

-
Kwon, M., R. N. Zakhidovna, K. B. Abdiazizovich, C. Jung and E.-J. Kil. 2024. First metagenomic analysis of virome in Uzbekistan honey bee (Apis mellifera): Investigating basic information on honey bee viruses. J. Invertebr. Pathol. 206: 108171.
[https://doi.org/10.1016/j.jip.2024.108171]

-
Letunic, I. and P. Bork. 2024. Interactive Tree of Life (iTOL) v6: recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 52(W1): W78-W82.
[https://doi.org/10.1093/nar/gkae268]

-
Li, N., C. Li, T. Hu, J. Li, H. Zhou, J. Ji, J. Wu, W. Kang, E. C. Holmes, W. Shi and S. Xu. 2023. Nationwide genomic surveillance reveals the prevalence and evolution of honeybee viruses in China. Microbiome 11(1): 6.
[https://doi.org/10.1186/s40168-022-01446-1]

-
Li, N., Y. Huang, W. Li and S. Xu. 2022. Virome analysis reveals diverse and divergent RNA viruses in wild insect pollinators in Beijing, China. Viruses 14(2): 227.
[https://doi.org/10.3390/v14020227]

-
Massart, S., M. Chiumenti, K. De Jonghe, R. Glover, A. Haegeman, I. Koloniuk, P. Komínek, J. Kreuze, D. Kutnjak, L. Lotos, F. Maclot, V. Maliogka, H. J. Maree, T. Olivier, A. Olmos, M. M. Pooggin, J.-S. Reynard, A. B. Ruiz-García, D. Safarova, P. H. H. Schneeberger, N. Sela, S. Turco, E. J. Vainio, E. Varallyay, E. Verdin, M. Westenberg, Y. Brostaux and T. Candresse. 2019. Virus detection by high-throughput sequencing of small RNAs: Large-scale performance testing of sequence analysis strategies. Phytopathology 109(3): 488-497.
[https://doi.org/10.1094/PHYTO-02-18-0067-R]

-
Minh, B. Q., H. A. Schmidt, O. Chernomor, D. Schrempf, M. D. Woodhams, A. Von Haeseler and R. Lanfear. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37(5): 1530-1534.
[https://doi.org/10.1093/molbev/msaa015]

-
Nannan, L., L. Huamiao, J. Yan, L. Xingan, L. Yang, W. Tianjiao, H. Jinming, N. Qingsheng and X. Xiumei.2022. Geometric morphology and population genomics provide insights into the adaptive evolution of Apis cerana in Changbai Mountain. BMC Genomics 23(1): 64.
[https://doi.org/10.1186/s12864-022-08298-x]

-
Nikulin, S. L., P. J. Hesketh-Best, D. A. Mckeown, M. Spivak and D. C. Schroeder. 2024. A semi-automated and high-throughput approach for the detection of honey bee viruses in bee samples. PLoS One 19(3): e0297623.
[https://doi.org/10.1371/journal.pone.0297623]

-
Pérez-Losada, M., M. Arenas, J. C. Galán, M. A. Bracho, J. Hillung, N. García-González and F. González-Candelas. 2020. High-throughput sequencing (HTS) for the analysis of viral populations. Infect. Genet. Evol. 80: 104208.
[https://doi.org/10.1016/j.meegid.2020.104208]

-
Roberts, J. M., K. B. Ireland, W. T. Tay and D. Paini. 2018. Honey bee-assisted surveillance for early plant virus detection. Ann. Appl. Biol. 173(3): 285-293.
[https://doi.org/10.1111/aab.12461]

-
Sbardellati, D. L. and R. L. Vannette. 2024. Targeted viromes and total metagenomes capture distinct components of bee gut phage communities. Microbiome 12(1): 155.
[https://doi.org/10.1186/s40168-024-01875-0]

-
Shi, M., X.-D. Lin, J.-H. Tian, L.-J. Chen, X. Chen, C.-X. Li, X.-C. Qin, J. Li, J.-P. Cao, J.-S. Eden, J. Buchmann, W. Wang, J. Xu, E. C. Holmes and Y.-Z. Zhang. 2016. Redefining the invertebrate RNA virosphere. Nature 540(7634): 539-543.
[https://doi.org/10.1038/nature20167]

-
Thu, H. T., N. T. Lien, P. T. Lanh, B. T. Duong, N. T. Hoa, M. H. Phuoc, P. H. Thai and D. Van Quyen. 2020. Genome analysis and phylogenetic characterization of two deformed wing virus strains from Apis cerana in Vietnam. PeerJ 8: e9911.
[https://doi.org/10.7717/peerj.9911]

-
Toplak, I., L. Šimenc, M. Pislak Ocepek and D. Bevk. 2020. Determination of genetically identical strains of four honeybee viruses in bumblebee positive samples. Viruses 12(11): 1310.
[https://doi.org/10.3390/v12111310]

-
Van Herzele, C., S. Coppens, N. Vereecke, S. Theuns, D. C. de Graaf and H. Nauwynck. 2024. New insights into honey bee viral and bacterial seasonal infection patterns using third-generation nanopore sequencing on honey bee haemolymph. Vet. Res. 55(1): 118.
[https://doi.org/10.1186/s13567-024-01382-y]

-
Villamor, D., T. Ho, M. Al Rwahnih, R. Martin and I. Tzanetakis. 2019. High throughput sequencing for plant virus detection and discovery. Phytopathology 109(5): 716-725.
[https://doi.org/10.1094/PHYTO-07-18-0257-RVW]

-
Vung, N. N., M.-L. Lee, M.-Y. Lee, H. K. Kim, E. J. Kang, J. E. Kim and Y.-S. Choi. 2017. Breeding and selection for resistance to sacbrood virus for Apis cerana. J. Apic. 32(4): 345-352.
[https://doi.org/10.17519/apiculture.2017.11.32.4.345]

-
Wolf, Y. I., D. Kazlauskas, J. Iranzo, A. Lucía-Sanz, J. H. Kuhn, M. Krupovic, V. V. Dolja and E. V. Koonin. 2018. Origins and evolution of the global RNA virome. MBio 9(6): 10.1128/mbio. 02329-02318.
[https://doi.org/10.1128/mbio.02329-18]

-
Zhang, C., S. Pokhrel, Z. Wu, X. Miao, Z. Y. Huang and W. Yang. 2019. Longevity, food consumption, and foraging performance of Apis cerana and Apis mellifera in mixed colonies. Apidologie 50: 153-162.
[https://doi.org/10.1007/s13592-018-0626-7]